Project
DANIO-CODE
Navigation
Downloads
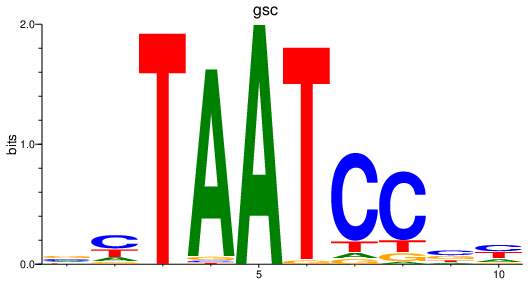
Results for gsc
Z-value: 1.01
Transcription factors associated with gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsc
|
ENSDARG00000059073 | goosecoid |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsc | dr10_dc_chr17_-_19325678_19325688 | 0.23 | 4.0e-01 | Click! |
Activity profile of gsc motif
Sorted Z-values of gsc motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gsc
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_26013873 | 4.87 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr17_-_32473712 | 2.58 |
ENSDART00000148455
ENSDART00000149885 |
grhl1
|
grainyhead-like transcription factor 1 |
| chr9_-_43178744 | 2.54 |
|
|
|
| chr22_+_14068525 | 2.48 |
ENSDART00000080329
ENSDART00000115383 |
he1a
|
hatching enzyme 1a |
| chr6_-_43094573 | 2.46 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr23_-_27226280 | 2.26 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr11_-_29316162 | 2.20 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr25_+_182037 | 2.04 |
ENSDART00000156469
|
RPS17
|
ribosomal protein S17 |
| chr19_-_32931355 | 2.00 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr14_-_36037883 | 1.90 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr21_-_39239757 | 1.83 |
|
|
|
| chr16_+_42925950 | 1.82 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr5_+_44316830 | 1.74 |
|
|
|
| chr9_+_38975508 | 1.73 |
|
|
|
| chr23_+_36832325 | 1.73 |
|
|
|
| chr5_+_59844830 | 1.72 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr5_+_59845054 | 1.70 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr15_-_15532980 | 1.58 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr23_-_27226387 | 1.57 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr16_+_25180444 | 1.56 |
ENSDART00000157346
ENSDART00000155032 |
si:ch211-261d7.3
|
si:ch211-261d7.3 |
| chr13_+_11305846 | 1.55 |
|
|
|
| chr9_+_26292183 | 1.55 |
|
|
|
| chr10_-_8029671 | 1.53 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr13_-_31491759 | 1.52 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr10_+_32160464 | 1.51 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr5_-_28025315 | 1.51 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr17_+_45931793 | 1.49 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr3_+_45375639 | 1.48 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr5_-_28025439 | 1.41 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr1_-_44476084 | 1.37 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr16_+_19730756 | 1.32 |
ENSDART00000112894
|
sp8b
|
sp8 transcription factor b |
| chr16_+_19730820 | 1.27 |
ENSDART00000079201
|
sp8b
|
sp8 transcription factor b |
| chr2_-_30071872 | 1.25 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr11_+_18823629 | 1.25 |
|
|
|
| chr10_-_37131530 | 1.24 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr18_+_45121016 | 1.22 |
ENSDART00000172328
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr13_-_50299821 | 1.19 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr23_-_5825236 | 1.17 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr11_+_15478585 | 1.16 |
ENSDART00000163289
ENSDART00000066033 |
gdf11
|
growth differentiation factor 11 |
| chr6_-_39278328 | 1.15 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr15_-_45229713 | 1.14 |
|
|
|
| chr17_-_6191539 | 1.13 |
ENSDART00000081707
|
ENSDARG00000058775
|
ENSDARG00000058775 |
| chr3_-_58739476 | 1.13 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr13_-_39821399 | 1.12 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr23_-_10202347 | 1.12 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr12_-_10001043 | 1.09 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr9_+_33167554 | 1.09 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr6_-_13215384 | 1.08 |
ENSDART00000112883
|
fmnl2b
|
formin-like 2b |
| chr14_+_34626233 | 1.00 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr19_-_42721857 | 0.98 |
ENSDART00000150919
ENSDART00000151034 |
si:ch211-191i18.2
|
si:ch211-191i18.2 |
| chr3_+_23573114 | 0.96 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr14_+_30390575 | 0.89 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr2_-_30071815 | 0.87 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr11_+_30048913 | 0.86 |
|
|
|
| chr13_-_33269297 | 0.85 |
|
|
|
| chr23_+_41295092 | 0.85 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr5_+_58824344 | 0.85 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr13_+_24417183 | 0.84 |
ENSDART00000177677
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr7_-_38288929 | 0.84 |
|
|
|
| chr11_-_40414239 | 0.83 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr13_-_50299643 | 0.83 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr7_-_49322226 | 0.82 |
ENSDART00000174161
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr2_-_30071993 | 0.80 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr23_+_14058972 | 0.80 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr2_-_42222069 | 0.79 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr12_+_5495284 | 0.77 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr21_-_91068 | 0.77 |
|
|
|
| chr4_-_14927871 | 0.76 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr5_-_56856570 | 0.76 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr23_-_4175790 | 0.76 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr23_-_10202175 | 0.75 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr23_+_6298911 | 0.75 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr22_-_26971466 | 0.73 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr24_+_37645966 | 0.73 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr11_+_25266623 | 0.71 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr8_+_14343513 | 0.69 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr3_-_58739399 | 0.69 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr2_-_42011586 | 0.68 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr14_+_47326080 | 0.67 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr3_+_45375698 | 0.67 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr18_-_43890514 | 0.65 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr11_+_19894390 | 0.63 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr17_+_45931756 | 0.59 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr5_-_21520111 | 0.56 |
|
|
|
| chr6_-_17699493 | 0.56 |
ENSDART00000154180
|
CR388029.1
|
ENSDARG00000097214 |
| chr6_+_54931721 | 0.54 |
|
|
|
| chr19_+_39100820 | 0.52 |
|
|
|
| chr17_-_15141309 | 0.50 |
ENSDART00000103405
|
gch1
|
GTP cyclohydrolase 1 |
| chr20_-_25537241 | 0.50 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr14_+_7393010 | 0.49 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr11_+_30049412 | 0.47 |
|
|
|
| chr6_-_35417797 | 0.47 |
ENSDART00000016586
|
rgs5a
|
regulator of G protein signaling 5a |
| chr5_+_69369417 | 0.47 |
|
|
|
| chr17_+_45932080 | 0.46 |
ENSDART00000157326
|
kif26ab
|
kinesin family member 26Ab |
| chr21_+_15660541 | 0.45 |
ENSDART00000101956
ENSDART00000026903 |
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr20_+_29685096 | 0.45 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr20_+_27088598 | 0.43 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr5_-_13890202 | 0.43 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr23_-_5825177 | 0.42 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr4_+_5370319 | 0.42 |
ENSDART00000163797
|
ENSDARG00000014395
|
ENSDARG00000014395 |
| chr19_-_42668754 | 0.41 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr3_-_16077831 | 0.40 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr10_+_13320935 | 0.39 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr7_-_21639345 | 0.38 |
ENSDART00000111066
|
epoa
|
erythropoietin a |
| chr21_-_14950265 | 0.38 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr8_+_18584936 | 0.38 |
ENSDART00000089161
|
ENSDARG00000062021
|
ENSDARG00000062021 |
| chr5_+_26217533 | 0.37 |
|
|
|
| chr11_-_40414131 | 0.37 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr5_-_24302467 | 0.37 |
|
|
|
| chr1_+_46400232 | 0.37 |
ENSDART00000036783
|
cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr1_-_45228793 | 0.33 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr20_+_32646617 | 0.33 |
|
|
|
| chr18_+_17765358 | 0.33 |
ENSDART00000137239
|
si:ch211-216l23.2
|
si:ch211-216l23.2 |
| chr16_-_22903823 | 0.33 |
|
|
|
| chr25_-_31970637 | 0.32 |
ENSDART00000006124
|
cops2
|
COP9 signalosome subunit 2 |
| chr7_-_50244215 | 0.32 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr7_+_66660781 | 0.31 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr14_-_29459799 | 0.31 |
ENSDART00000005568
|
pdlim3b
|
PDZ and LIM domain 3b |
| chr5_+_42472789 | 0.30 |
ENSDART00000026020
|
wdr54
|
WD repeat domain 54 |
| chr24_-_16760535 | 0.30 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr11_+_2437479 | 0.30 |
ENSDART00000130886
|
NABP2
|
nucleic acid binding protein 2 |
| chr23_+_5631757 | 0.29 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr24_+_9740818 | 0.28 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr12_-_10667591 | 0.27 |
ENSDART00000164038
|
ENSDARG00000101171
|
ENSDARG00000101171 |
| chr19_+_19224936 | 0.26 |
ENSDART00000171103
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr8_+_28493939 | 0.25 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr13_+_11305968 | 0.24 |
|
|
|
| chr15_+_30580253 | 0.24 |
|
|
|
| chr8_+_28419432 | 0.24 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr21_-_41994224 | 0.23 |
ENSDART00000092821
ENSDART00000165743 ENSDART00000075740 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr9_-_28588288 | 0.21 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr3_-_5367142 | 0.20 |
ENSDART00000098927
|
txn2
|
thioredoxin 2 |
| chr18_+_7680416 | 0.19 |
ENSDART00000132369
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr19_+_19225228 | 0.19 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_59845172 | 0.18 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr2_-_22149297 | 0.17 |
|
|
|
| chr5_+_25346893 | 0.17 |
|
|
|
| chr5_+_63716392 | 0.16 |
ENSDART00000015940
|
edf1
|
endothelial differentiation-related factor 1 |
| chr5_-_56856527 | 0.16 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr7_+_30698873 | 0.16 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr25_+_15983777 | 0.15 |
ENSDART00000165598
ENSDART00000061753 |
far1
|
fatty acyl CoA reductase 1 |
| chr25_-_22791839 | 0.15 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr19_-_25842461 | 0.14 |
|
|
|
| chr19_+_19224736 | 0.13 |
ENSDART00000158228
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr23_-_17583239 | 0.12 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr3_-_30810423 | 0.12 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr23_-_28368224 | 0.12 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr6_-_39278397 | 0.11 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr8_+_36951655 | 0.10 |
|
|
|
| chr7_+_52612436 | 0.10 |
ENSDART00000109973
|
tp53bp1
|
tumor protein p53 binding protein, 1 |
| chr14_-_46564205 | 0.10 |
ENSDART00000111410
|
ganab
|
glucosidase, alpha; neutral AB |
| chr17_-_8498935 | 0.10 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr17_-_20959288 | 0.10 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr17_-_33462243 | 0.09 |
ENSDART00000131807
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr20_-_18836593 | 0.09 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr6_-_13215294 | 0.08 |
ENSDART00000112883
|
fmnl2b
|
formin-like 2b |
| chr4_+_14928134 | 0.07 |
ENSDART00000019647
|
psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr24_+_3447129 | 0.06 |
ENSDART00000134598
|
wdr37
|
WD repeat domain 37 |
| chr3_+_23573053 | 0.06 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr7_+_41141295 | 0.05 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr24_-_16760393 | 0.05 |
ENSDART00000149706
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr20_+_29685331 | 0.04 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr17_+_45932212 | 0.02 |
ENSDART00000157326
|
kif26ab
|
kinesin family member 26Ab |
| chr5_+_18941935 | 0.02 |
ENSDART00000163064
|
ube3b
|
ubiquitin protein ligase E3B |
| chr6_+_30385605 | 0.01 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr19_+_19225148 | 0.01 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr2_+_36845714 | 0.01 |
ENSDART00000140474
|
nrd1a
|
nardilysin a (N-arginine dibasic convertase) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 0.8 | 2.5 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.6 | 2.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.6 | 1.9 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.5 | 3.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.4 | 2.6 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.4 | 1.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 1.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.4 | 2.5 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.3 | 3.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 1.9 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 1.5 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.2 | 2.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 2.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 1.6 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 | 1.5 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.7 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.6 | GO:1904407 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 0.7 | GO:0097345 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 1.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.1 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 1.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.4 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.2 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.9 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 1.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.2 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) regulation of postsynaptic membrane potential(GO:0060078) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.7 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 1.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 5.8 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.5 | 4.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 1.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 2.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.8 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.4 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 2.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 2.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.8 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |