Project
DANIO-CODE
Navigation
Downloads
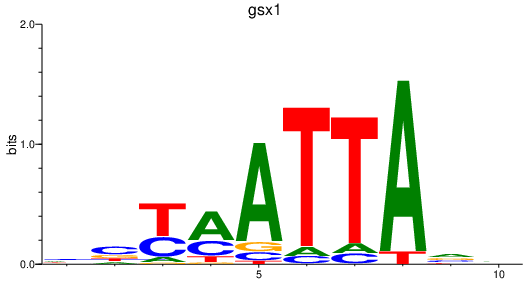
Results for gsx1
Z-value: 1.06
Transcription factors associated with gsx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsx1
|
ENSDARG00000035735 | GS homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsx1 | dr10_dc_chr5_-_67233396_67233400 | 0.64 | 7.2e-03 | Click! |
Activity profile of gsx1 motif
Sorted Z-values of gsx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_68410884 | 4.47 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr6_+_56157608 | 2.95 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr12_+_3043444 | 2.57 |
ENSDART00000149427
|
sgca
|
sarcoglycan, alpha |
| chr14_-_26238386 | 2.23 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr14_+_46419051 | 2.16 |
ENSDART00000112377
|
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr19_-_24971633 | 2.07 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr11_-_30387031 | 2.02 |
|
|
|
| chr2_+_33343287 | 1.99 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr8_-_23172938 | 1.98 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr21_+_28408329 | 1.79 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr13_+_27102377 | 1.77 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr9_-_32942783 | 1.73 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr21_+_25199691 | 1.71 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr24_-_6048914 | 1.68 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr21_-_23074223 | 1.65 |
ENSDART00000147896
|
usp28
|
ubiquitin specific peptidase 28 |
| chr6_-_2294751 | 1.65 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr11_+_37639045 | 1.62 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr8_+_30443651 | 1.61 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr6_-_23193752 | 1.59 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr22_-_15567180 | 1.58 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr1_+_46487300 | 1.53 |
ENSDART00000158432
ENSDART00000074450 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr13_+_7575753 | 1.52 |
|
|
|
| chr1_-_58580939 | 1.50 |
ENSDART00000158011
|
col5a3b
|
collagen, type V, alpha 3b |
| chr8_+_46651079 | 1.48 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr21_+_26684617 | 1.46 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr7_+_34018862 | 1.46 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr19_-_31815128 | 1.44 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr1_+_32935645 | 1.43 |
ENSDART00000170832
|
arl13b
|
ADP-ribosylation factor-like 13b |
| chr3_-_29768364 | 1.42 |
ENSDART00000020311
|
rpl27
|
ribosomal protein L27 |
| chr16_-_17439735 | 1.39 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr18_+_730277 | 1.39 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr18_-_3056732 | 1.38 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr23_-_12310778 | 1.37 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr11_+_10925475 | 1.36 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr20_-_9107294 | 1.35 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr20_+_19613133 | 1.35 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr22_+_223797 | 1.30 |
|
|
|
| chr8_+_21321765 | 1.26 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr16_+_46145286 | 1.24 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr16_+_43441389 | 1.19 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr21_-_17259886 | 1.17 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr25_+_14411153 | 1.17 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr5_-_13872895 | 1.16 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr11_-_2107054 | 1.16 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr8_+_23716843 | 1.14 |
ENSDART00000136547
|
rpl10a
|
ribosomal protein L10a |
| chr9_-_20562293 | 1.14 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr7_-_46510632 | 1.14 |
|
|
|
| chr20_-_40823307 | 1.13 |
ENSDART00000061261
|
cx43
|
connexin 43 |
| chr8_-_23591082 | 1.13 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr5_+_2384929 | 1.10 |
|
|
|
| chr25_+_5856023 | 1.09 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr5_-_32687349 | 1.09 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr12_+_24221087 | 1.05 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr16_-_21238288 | 1.03 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr24_+_35500964 | 1.03 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr2_-_24113073 | 1.03 |
ENSDART00000148685
|
xirp1
|
xin actin binding repeat containing 1 |
| chr15_-_1858350 | 1.03 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr7_+_20251345 | 1.03 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr9_+_19087329 | 1.00 |
ENSDART00000110457
|
crfb1
|
cytokine receptor family member b1 |
| chr7_-_17129072 | 1.00 |
|
|
|
| chr6_-_46939315 | 0.99 |
ENSDART00000129301
ENSDART00000037875 |
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr16_-_28943421 | 0.96 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr24_+_37752791 | 0.93 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr16_-_43441084 | 0.92 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr10_+_2772113 | 0.90 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| KN150623v1_+_577 | 0.89 |
|
|
|
| chr14_-_4166292 | 0.88 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr24_-_2278409 | 0.87 |
|
|
|
| chr25_+_1747864 | 0.84 |
|
|
|
| chr8_+_17132156 | 0.84 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr1_-_25600988 | 0.83 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr16_+_13928844 | 0.80 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr10_+_2952793 | 0.80 |
|
|
|
| chr13_+_29108288 | 0.79 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr14_+_160149 | 0.79 |
ENSDART00000158405
ENSDART00000158072 |
gpc2
|
glypican 2 |
| chr2_-_36058327 | 0.79 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr18_-_19467100 | 0.77 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr9_+_36316158 | 0.74 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr15_-_23441268 | 0.73 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr9_-_19154264 | 0.73 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr6_+_53349584 | 0.72 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr17_+_1799209 | 0.70 |
ENSDART00000112183
|
cep170b
|
centrosomal protein 170B |
| chr1_-_54561569 | 0.69 |
ENSDART00000110818
|
ENSDARG00000036281
|
ENSDARG00000036281 |
| chr7_+_16854284 | 0.67 |
ENSDART00000013409
|
prmt3
|
protein arginine methyltransferase 3 |
| chr6_-_57542101 | 0.66 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr20_+_602264 | 0.65 |
|
|
|
| chr24_-_36227334 | 0.65 |
|
|
|
| chr23_+_13106555 | 0.65 |
|
|
|
| chr9_-_25437880 | 0.65 |
|
|
|
| chr15_-_45446153 | 0.64 |
ENSDART00000100332
|
fgf12b
|
fibroblast growth factor 12b |
| chr8_-_18502159 | 0.64 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr16_+_51319421 | 0.64 |
|
|
|
| chr21_+_7737034 | 0.64 |
|
|
|
| chr23_+_11350727 | 0.64 |
|
|
|
| chr1_-_50215233 | 0.62 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr12_-_31609517 | 0.60 |
ENSDART00000080173
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr9_-_41072329 | 0.59 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr18_-_34573597 | 0.58 |
ENSDART00000021880
|
ssr3
|
signal sequence receptor, gamma |
| chr6_-_23193873 | 0.58 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr25_-_5883620 | 0.57 |
ENSDART00000136054
|
snx22
|
sorting nexin 22 |
| chr11_-_34258956 | 0.55 |
ENSDART00000114004
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr24_+_9272045 | 0.54 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr3_+_47412661 | 0.54 |
ENSDART00000155648
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr3_-_29739166 | 0.54 |
ENSDART00000147048
|
rpl3
|
ribosomal protein L3 |
| chr9_-_39190534 | 0.53 |
|
|
|
| chr24_+_33697332 | 0.53 |
|
|
|
| chr25_+_16593169 | 0.53 |
ENSDART00000073416
|
cecr1a
|
cat eye syndrome chromosome region, candidate 1a |
| chr7_+_20377678 | 0.52 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr20_-_46458839 | 0.52 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr20_+_1724609 | 0.52 |
|
|
|
| chr1_+_46487378 | 0.51 |
ENSDART00000137448
|
morc3b
|
MORC family CW-type zinc finger 3b |
| chr5_-_71277652 | 0.50 |
|
|
|
| chr19_-_44064157 | 0.48 |
ENSDART00000144058
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr5_-_13738619 | 0.48 |
ENSDART00000090788
|
npffr1l2
|
neuropeptide FF receptor 1 like 2 |
| chr1_+_44044807 | 0.47 |
ENSDART00000133926
ENSDART00000049701 |
p2rx3b
|
purinergic receptor P2X, ligand-gated ion channel, 3b |
| chr24_-_2453987 | 0.46 |
ENSDART00000093331
|
rreb1a
|
ras responsive element binding protein 1a |
| chr17_+_25425967 | 0.45 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr5_-_33156615 | 0.43 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr19_-_30923513 | 0.42 |
ENSDART00000088760
|
bag6l
|
BCL2-associated athanogene 6, like |
| chr20_-_23526954 | 0.41 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr15_-_16047125 | 0.41 |
ENSDART00000166583
|
synrg
|
synergin, gamma |
| KN150623v1_+_422 | 0.41 |
|
|
|
| chr18_-_43890514 | 0.40 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr8_+_25015374 | 0.39 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr21_-_27584637 | 0.38 |
|
|
|
| chr9_-_43736549 | 0.37 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr9_+_36023553 | 0.37 |
ENSDART00000122169
|
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr2_+_29507819 | 0.36 |
|
|
|
| chr24_+_39750685 | 0.34 |
ENSDART00000133284
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr21_+_26684728 | 0.34 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr24_+_16402613 | 0.33 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_+_53384116 | 0.33 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr5_+_32687452 | 0.32 |
ENSDART00000123210
|
med22
|
mediator complex subunit 22 |
| chr6_-_2294717 | 0.31 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr5_+_65754237 | 0.31 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr24_+_40292617 | 0.31 |
|
|
|
| chr12_-_268910 | 0.29 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr11_-_460003 | 0.28 |
ENSDART00000093148
|
isy1
|
ISY1 splicing factor homolog |
| chr16_-_16274403 | 0.28 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr3_+_53062322 | 0.28 |
ENSDART00000011780
|
xab2
|
XPA binding protein 2 |
| chr23_+_31986806 | 0.28 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr6_-_46939390 | 0.25 |
ENSDART00000129301
ENSDART00000037875 |
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr2_-_19532470 | 0.25 |
ENSDART00000166881
|
BX323588.1
|
ENSDARG00000102520 |
| chr8_+_25015325 | 0.25 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr13_+_4096622 | 0.25 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr19_-_30923133 | 0.25 |
ENSDART00000135128
|
bag6l
|
BCL2-associated athanogene 6, like |
| chr23_-_33017920 | 0.25 |
|
|
|
| chr5_+_37240702 | 0.24 |
ENSDART00000097738
|
panx1b
|
pannexin 1b |
| chr25_+_32348377 | 0.24 |
ENSDART00000162188
|
ETFA
|
electron transfer flavoprotein alpha subunit |
| chr3_-_29932835 | 0.24 |
|
|
|
| chr1_-_8439215 | 0.23 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr13_+_7575720 | 0.22 |
|
|
|
| chr3_-_16569378 | 0.22 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr11_+_1524965 | 0.21 |
ENSDART00000040577
|
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr16_+_17808913 | 0.19 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr13_+_33173790 | 0.18 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr20_-_23527004 | 0.17 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_-_11365575 | 0.17 |
ENSDART00000140217
|
prpf4bb
|
pre-mRNA processing factor 4Bb |
| chr21_-_11877525 | 0.17 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr18_+_15872911 | 0.14 |
ENSDART00000141800
|
eea1
|
early endosome antigen 1 |
| chr23_+_20779487 | 0.13 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr4_+_62081023 | 0.11 |
ENSDART00000160964
|
si:dkey-92j16.2
|
si:dkey-92j16.2 |
| chr22_-_864745 | 0.11 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr2_-_14833381 | 0.10 |
|
|
|
| chr17_-_7990960 | 0.09 |
|
|
|
| chr10_+_6925975 | 0.09 |
|
|
|
| chr16_-_53008678 | 0.09 |
|
|
|
| chr20_+_5511996 | 0.09 |
|
|
|
| chr10_+_8239066 | 0.08 |
ENSDART00000176831
|
skiv2l2
|
superkiller viralicidic activity 2-like 2 |
| chr20_-_43281081 | 0.07 |
|
|
|
| chr4_+_5789935 | 0.06 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr23_-_31986482 | 0.06 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr7_-_7530898 | 0.06 |
ENSDART00000113131
|
intu
|
inturned planar cell polarity protein |
| chr2_-_14833347 | 0.05 |
|
|
|
| chr3_+_27655753 | 0.05 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr1_-_28335293 | 0.04 |
ENSDART00000075546
|
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr24_-_37752716 | 0.03 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr14_+_23419894 | 0.01 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr2_-_55565099 | 0.01 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr15_-_16241341 | 0.00 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.5 | 2.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.5 | 1.4 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.3 | 1.3 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 1.8 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 0.7 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.2 | 0.6 | GO:0032329 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.8 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 2.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.4 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.6 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.5 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 3.0 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.1 | 1.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.5 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) respiratory tube development(GO:0030323) lung development(GO:0030324) synaptonemal complex organization(GO:0070193) |
| 0.1 | 2.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.4 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.6 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 2.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 2.9 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.2 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 2.6 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.0 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.4 | 2.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.3 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 2.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 3.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0051192 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 3.8 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 1.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.6 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 2.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 6.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |