Project
DANIO-CODE
Navigation
Downloads
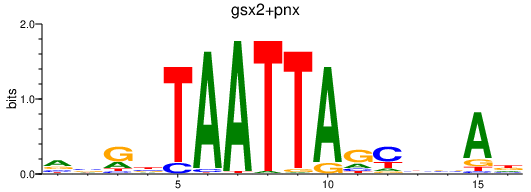
Results for gsx2+pnx
Z-value: 0.83
Transcription factors associated with gsx2+pnx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pnx
|
ENSDARG00000025899 | posterior neuron-specific homeobox |
|
gsx2
|
ENSDARG00000043322 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pnx | dr10_dc_chr10_+_2772113_2772118 | 0.62 | 1.0e-02 | Click! |
| gsx2 | dr10_dc_chr20_-_22584941_22584953 | 0.59 | 1.5e-02 | Click! |
Activity profile of gsx2+pnx motif
Sorted Z-values of gsx2+pnx motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx2+pnx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_35967525 | 2.84 |
ENSDART00000176971
|
CABZ01037120.1
|
ENSDARG00000106725 |
| chr20_-_36984259 | 2.36 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr13_+_35213326 | 2.00 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr9_-_30437160 | 1.90 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr12_-_4497094 | 1.84 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr14_+_6122948 | 1.81 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr17_-_35934175 | 1.79 |
ENSDART00000110040
ENSDART00000137525 |
sox11a
AL929378.1
|
SRY (sex determining region Y)-box 11a ENSDARG00000092907 |
| chr14_-_14337584 | 1.77 |
ENSDART00000167119
ENSDART00000168027 ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| chr3_-_23513177 | 1.71 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr14_-_40022693 | 1.70 |
ENSDART00000149443
|
pcdh19
|
protocadherin 19 |
| chr25_+_30699938 | 1.66 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr11_-_15162109 | 1.65 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr11_-_29910947 | 1.63 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr7_+_56405531 | 1.62 |
|
|
|
| chr25_+_31457309 | 1.58 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr19_+_17451381 | 1.58 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr1_-_57300743 | 1.58 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr12_+_27032862 | 1.48 |
|
|
|
| chr19_-_7576069 | 1.40 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr21_+_20512024 | 1.40 |
ENSDART00000126005
|
efna5a
|
ephrin-A5a |
| chr6_-_23193752 | 1.40 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr20_-_9107294 | 1.38 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr15_-_4537178 | 1.36 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr14_-_4038642 | 1.36 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr17_+_23278879 | 1.30 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr12_+_20230575 | 1.26 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr23_-_12310778 | 1.26 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr23_+_19887319 | 1.26 |
ENSDART00000139192
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr17_+_134921 | 1.22 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr12_-_27032151 | 1.21 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr11_-_446028 | 1.16 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr2_-_24898678 | 1.15 |
ENSDART00000145145
|
cnn2
|
calponin 2 |
| chr1_-_58580939 | 1.13 |
ENSDART00000158011
|
col5a3b
|
collagen, type V, alpha 3b |
| chr17_+_135165 | 1.13 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr11_-_6442836 | 1.11 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr25_-_13737344 | 1.10 |
|
|
|
| chr6_-_7563110 | 1.08 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr18_-_46260401 | 1.08 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| KN149726v1_+_1094 | 1.07 |
|
|
|
| chr7_+_30516734 | 1.07 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr9_+_36092350 | 1.05 |
ENSDART00000005086
|
atp1a1b
|
ATPase, Na+/K+ transporting, alpha 1b polypeptide |
| chr23_+_44137593 | 1.05 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr20_+_545574 | 1.05 |
ENSDART00000152453
|
dse
|
dermatan sulfate epimerase |
| chr3_-_23277066 | 1.04 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr25_+_34515221 | 1.04 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr21_-_3548719 | 1.03 |
ENSDART00000137844
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr24_+_37752791 | 1.00 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr4_+_73490746 | 0.97 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr23_+_20183765 | 0.97 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr23_+_28656263 | 0.96 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr8_-_15091823 | 0.96 |
ENSDART00000142358
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr3_+_13114740 | 0.96 |
ENSDART00000162724
|
ENSDARG00000074231
|
ENSDARG00000074231 |
| chr10_+_32160464 | 0.95 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr19_+_21783429 | 0.94 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr8_+_7740132 | 0.93 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr10_+_2952793 | 0.93 |
|
|
|
| chr7_-_17129072 | 0.93 |
|
|
|
| chr23_+_27052594 | 0.92 |
ENSDART00000109712
ENSDART00000018654 |
rnd1b
|
Rho family GTPase 1b |
| chr6_+_34528904 | 0.90 |
ENSDART00000161249
ENSDART00000166715 ENSDART00000161502 |
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr3_-_60866373 | 0.89 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr12_-_28248133 | 0.89 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr13_-_25502297 | 0.88 |
ENSDART00000077627
ENSDART00000139237 |
ret
|
ret proto-oncogene receptor tyrosine kinase |
| chr4_+_292818 | 0.87 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr15_-_14616083 | 0.86 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr15_-_35394535 | 0.86 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr1_+_55077656 | 0.86 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr3_-_38642067 | 0.85 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr20_+_10502210 | 0.84 |
ENSDART00000155888
|
AL845550.1
|
ENSDARG00000097342 |
| chr15_-_1858350 | 0.84 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr11_+_37639045 | 0.84 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr13_-_49695322 | 0.84 |
|
|
|
| chr20_-_15734333 | 0.83 |
ENSDART00000176999
ENSDART00000179202 |
CABZ01015860.1
|
ENSDARG00000108793 |
| chr7_-_23775835 | 0.82 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr1_-_50215233 | 0.79 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| KN149830v1_+_5168 | 0.79 |
|
|
|
| chr19_+_48555386 | 0.79 |
|
|
|
| chr16_+_51319421 | 0.78 |
|
|
|
| chr22_+_16282153 | 0.78 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr16_+_1302842 | 0.78 |
|
|
|
| chr24_-_11768200 | 0.77 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_-_15584548 | 0.75 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr6_+_6767424 | 0.75 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr2_+_33399405 | 0.75 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr5_+_39885307 | 0.73 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr1_+_6833994 | 0.71 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr8_-_31098193 | 0.70 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr19_-_22956977 | 0.69 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr23_-_31585883 | 0.67 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr24_-_6048914 | 0.65 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr20_+_29307039 | 0.64 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr4_+_75129077 | 0.64 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr8_+_1760968 | 0.64 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr20_+_29307142 | 0.64 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr9_-_41608298 | 0.64 |
|
|
|
| chr13_+_18414806 | 0.63 |
ENSDART00000110197
|
zgc:154058
|
zgc:154058 |
| chr3_+_59491893 | 0.62 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr7_-_67906887 | 0.62 |
ENSDART00000171943
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr21_+_6829161 | 0.61 |
ENSDART00000037265
|
olfm1b
|
olfactomedin 1b |
| chr1_+_49894685 | 0.61 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr21_+_25496316 | 0.60 |
|
|
|
| chr6_-_30671802 | 0.59 |
ENSDART00000065215
|
lurap1
|
leucine rich adaptor protein 1 |
| chr1_-_518543 | 0.58 |
ENSDART00000138032
|
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr1_-_661529 | 0.58 |
ENSDART00000166786
ENSDART00000170483 |
appa
|
amyloid beta (A4) precursor protein a |
| chr6_-_39346614 | 0.58 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr8_-_19166630 | 0.58 |
|
|
|
| chr25_+_4909062 | 0.57 |
ENSDART00000170049
|
parvb
|
parvin, beta |
| chr11_+_1495509 | 0.57 |
ENSDART00000139079
|
srsf6b
|
serine/arginine-rich splicing factor 6b |
| chr24_+_35500964 | 0.57 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr11_-_41241693 | 0.57 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr14_+_5078937 | 0.56 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr7_-_19390325 | 0.55 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr8_+_52633069 | 0.54 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr11_+_43924640 | 0.54 |
ENSDART00000179206
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr6_+_24299180 | 0.53 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr1_-_57301027 | 0.53 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr10_-_26782374 | 0.53 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr14_-_3951077 | 0.53 |
|
|
|
| chr6_-_23193873 | 0.52 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr25_-_2386615 | 0.52 |
|
|
|
| chr19_+_32679320 | 0.52 |
|
|
|
| chr5_-_30015572 | 0.52 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr23_+_36016450 | 0.51 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr19_-_22957035 | 0.51 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr24_+_33697332 | 0.51 |
|
|
|
| chr3_+_13487523 | 0.51 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr4_+_22759177 | 0.51 |
ENSDART00000146272
|
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr19_-_3208938 | 0.51 |
|
|
|
| chr5_-_27958815 | 0.51 |
ENSDART00000177313
|
si:ch73-195i19.3
|
si:ch73-195i19.3 |
| chr3_-_60985683 | 0.51 |
ENSDART00000133091
|
pvalb5
|
parvalbumin 5 |
| chr5_-_23649439 | 0.50 |
|
|
|
| chr15_-_45446153 | 0.50 |
ENSDART00000100332
|
fgf12b
|
fibroblast growth factor 12b |
| chr23_-_10692678 | 0.49 |
|
|
|
| chr19_+_32679258 | 0.49 |
|
|
|
| chr4_-_935448 | 0.49 |
ENSDART00000171855
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr3_-_31392769 | 0.48 |
|
|
|
| chr11_+_18710724 | 0.48 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr15_+_34007335 | 0.47 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr21_-_26453406 | 0.47 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr23_-_36198253 | 0.46 |
|
|
|
| chr14_-_9407573 | 0.46 |
ENSDART00000166739
|
NRK
|
si:zfos-2326c3.2 |
| chr20_-_46458839 | 0.46 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| KN150127v1_+_2671 | 0.46 |
|
|
|
| chr18_-_43890514 | 0.46 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr9_+_36316158 | 0.45 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr15_-_18110169 | 0.44 |
|
|
|
| chr20_+_35479511 | 0.44 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr23_+_17220363 | 0.43 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr21_-_45550182 | 0.43 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr20_+_26981663 | 0.43 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr11_-_18068313 | 0.42 |
|
|
|
| chr24_+_40292617 | 0.42 |
|
|
|
| chr11_+_38013238 | 0.42 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr5_-_63031745 | 0.42 |
|
|
|
| chr13_+_47175 | 0.42 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr25_-_35530474 | 0.42 |
|
|
|
| chr20_+_25326042 | 0.40 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
| chr13_-_44492897 | 0.40 |
|
|
|
| chr5_+_65754237 | 0.40 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr5_-_68876416 | 0.40 |
ENSDART00000170885
|
eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha |
| chr19_+_48555339 | 0.40 |
|
|
|
| chr8_-_22252973 | 0.40 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| KN149830v1_-_23361 | 0.38 |
|
|
|
| chr23_-_46268313 | 0.38 |
ENSDART00000170417
ENSDART00000168352 |
CABZ01102528.1
|
ENSDARG00000098990 |
| chr21_+_45802046 | 0.37 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr17_+_6295257 | 0.37 |
ENSDART00000064694
|
TATDN3
|
TatD DNase domain containing 3 |
| chr20_-_23345739 | 0.37 |
|
|
|
| chr16_+_13928376 | 0.37 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr14_-_6831308 | 0.37 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr24_-_24859334 | 0.36 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr13_-_51409167 | 0.36 |
ENSDART00000174550
|
CABZ01090417.1
|
ENSDARG00000108080 |
| chr4_+_11459212 | 0.36 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr12_-_41128584 | 0.36 |
|
|
|
| chr17_-_37247505 | 0.35 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr11_-_40464213 | 0.35 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr3_+_22392439 | 0.35 |
|
|
|
| chr11_-_42263721 | 0.34 |
ENSDART00000130573
|
atp6ap1la
|
ATPase, H+ transporting, lysosomal accessory protein 1-like a |
| chr6_-_40886902 | 0.34 |
ENSDART00000017968
ENSDART00000154100 |
sirt4
|
sirtuin 4 |
| chr19_+_21783111 | 0.34 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr9_-_30765708 | 0.34 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr6_-_57542101 | 0.33 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr21_-_45550252 | 0.33 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr16_-_43029548 | 0.33 |
ENSDART00000057305
|
thbs3a
|
thrombospondin 3a |
| chr8_+_25015374 | 0.33 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr4_+_15072119 | 0.33 |
|
|
|
| chr1_+_127240 | 0.32 |
ENSDART00000122230
|
f7i
|
coagulation factor VIIi |
| chr13_-_40600924 | 0.32 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr17_+_135261 | 0.32 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr2_-_27065203 | 0.32 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr20_+_35479428 | 0.32 |
ENSDART00000159483
ENSDART00000031091 |
BX511024.1
vsnl1a
|
ENSDARG00000104812 visinin-like 1a |
| chr3_+_27667194 | 0.32 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr23_+_20779487 | 0.32 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr2_+_50873893 | 0.32 |
|
|
|
| chr3_-_30494171 | 0.32 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr11_+_26238579 | 0.31 |
ENSDART00000165097
|
unm_sa1614
|
un-named sa1614 |
| chr20_-_15029379 | 0.31 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr10_-_41478085 | 0.31 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr6_+_40924749 | 0.31 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_+_9441851 | 0.31 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr6_-_39201300 | 0.30 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr20_+_17899547 | 0.30 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.3 | 0.9 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.3 | 0.9 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 1.7 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 0.6 | GO:0097702 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.2 | 2.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 1.2 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.2 | 0.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.7 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.2 | 0.9 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.4 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.1 | 1.2 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 1.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.7 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 2.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.0 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.0 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.1 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.0 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.3 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.7 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.3 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 1.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.0 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.9 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 0.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 1.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 1.3 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.6 | GO:0042645 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.1 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.6 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 1.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 1.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.6 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 2.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.9 | GO:0019199 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) transmembrane receptor protein kinase activity(GO:0019199) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.1 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.5 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.0 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |