Project
DANIO-CODE
Navigation
Downloads
Results for hbp1
Z-value: 0.49
Transcription factors associated with hbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hbp1
|
ENSDARG00000028517 | HMG-box transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hbp1 | dr10_dc_chr25_-_3345176_3345205 | 0.64 | 7.4e-03 | Click! |
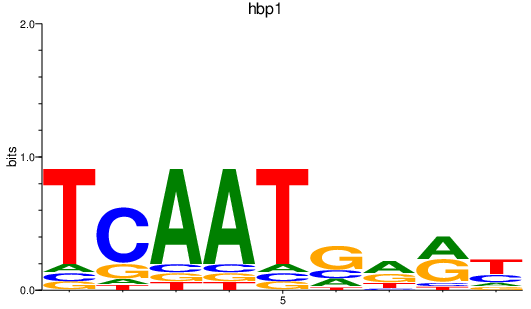
Activity profile of hbp1 motif
Sorted Z-values of hbp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hbp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_31606213 | 1.10 |
ENSDART00000126873
|
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr7_-_8128948 | 1.03 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr19_-_43037791 | 0.99 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr24_+_4341242 | 0.90 |
ENSDART00000133360
|
ccny
|
cyclin Y |
| chr6_+_23787804 | 0.79 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr14_+_5852295 | 0.68 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr2_+_9263844 | 0.65 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr9_+_30767182 | 0.65 |
ENSDART00000139811
|
commd6
|
COMM domain containing 6 |
| chr25_+_558191 | 0.63 |
ENSDART00000126863
ENSDART00000166012 |
nell2a
|
neural EGFL like 2a |
| chr20_-_44598129 | 0.59 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr20_+_26981663 | 0.55 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr7_-_25426346 | 0.54 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr15_+_16950266 | 0.54 |
ENSDART00000049196
|
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr6_-_24003717 | 0.53 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr19_+_43843780 | 0.51 |
ENSDART00000151571
|
ahdc1
|
AT hook, DNA binding motif, containing 1 |
| chr17_-_26893412 | 0.51 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr13_-_12513113 | 0.48 |
|
|
|
| chr14_-_2278018 | 0.48 |
ENSDART00000125674
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr6_+_60190849 | 0.46 |
|
|
|
| chr12_-_48948928 | 0.46 |
ENSDART00000163734
|
rgrb
|
retinal G protein coupled receptor b |
| chr3_+_43010408 | 0.45 |
ENSDART00000169061
|
CU138533.1
|
ENSDARG00000099842 |
| chr17_+_37306052 | 0.45 |
ENSDART00000134000
|
tmem62
|
transmembrane protein 62 |
| chr7_-_23751107 | 0.42 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr8_+_48624273 | 0.41 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr24_-_6868340 | 0.40 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr6_+_54931721 | 0.39 |
|
|
|
| chr14_-_34172522 | 0.38 |
ENSDART00000140368
|
si:ch211-232m8.3
|
si:ch211-232m8.3 |
| chr19_+_10912601 | 0.37 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr13_-_49546922 | 0.36 |
|
|
|
| chr4_+_9027862 | 0.36 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr9_-_34068225 | 0.36 |
ENSDART00000088441
ENSDART00000150035 |
si:ch73-147f11.1
|
si:ch73-147f11.1 |
| chr11_+_24640015 | 0.34 |
ENSDART00000006038
|
shisa4
|
shisa family member 4 |
| chr7_+_73466424 | 0.33 |
ENSDART00000123429
|
si:dkey-46i9.6
|
si:dkey-46i9.6 |
| chr6_-_40724581 | 0.33 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr25_-_34677352 | 0.33 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr23_+_23255489 | 0.31 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr16_+_5625552 | 0.31 |
|
|
|
| chr1_+_39545535 | 0.30 |
ENSDART00000045697
|
zgc:56493
|
zgc:56493 |
| chr18_+_17622557 | 0.30 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr18_+_11747188 | 0.29 |
ENSDART00000051105
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr11_-_25181234 | 0.29 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr24_+_25872051 | 0.28 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr18_-_19114558 | 0.28 |
ENSDART00000177621
|
dennd4a
|
DENN/MADD domain containing 4A |
| chr15_+_32439470 | 0.26 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr16_-_54638972 | 0.25 |
|
|
|
| chr1_-_32647357 | 0.25 |
ENSDART00000136383
|
cd99
|
CD99 molecule |
| chr19_-_32900108 | 0.25 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr17_-_45036982 | 0.24 |
ENSDART00000155043
|
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr23_-_41002187 | 0.24 |
ENSDART00000061037
|
ENSDARG00000041644
|
ENSDARG00000041644 |
| chr14_-_17000025 | 0.24 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr7_+_27420387 | 0.23 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr12_-_17079132 | 0.22 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr3_-_23277066 | 0.22 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_-_12584179 | 0.22 |
|
|
|
| chr10_-_20567013 | 0.22 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr22_+_29694605 | 0.22 |
|
|
|
| chr11_-_3592968 | 0.21 |
ENSDART00000085855
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr2_-_19532470 | 0.21 |
ENSDART00000166881
|
BX323588.1
|
ENSDARG00000102520 |
| chr18_-_39797167 | 0.21 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr14_-_34172619 | 0.20 |
ENSDART00000147380
|
ENSDARG00000092889
|
ENSDARG00000092889 |
| chr13_-_22713071 | 0.19 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr8_+_50498398 | 0.18 |
|
|
|
| chr14_+_5852428 | 0.17 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr16_+_22114192 | 0.17 |
ENSDART00000167919
|
znf687a
|
zinc finger protein 687a |
| chr7_+_28630661 | 0.16 |
ENSDART00000113998
|
dok4
|
docking protein 4 |
| chr10_+_35479226 | 0.16 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr19_+_46647893 | 0.15 |
ENSDART00000162785
ENSDART00000171251 ENSDART00000164938 |
mapk15
|
mitogen-activated protein kinase 15 |
| chr1_-_9364331 | 0.13 |
ENSDART00000054835
ENSDART00000125358 |
rnf175
|
ring finger protein 175 |
| chr1_-_54366578 | 0.12 |
ENSDART00000170001
|
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr6_-_42143595 | 0.11 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr13_+_13875110 | 0.11 |
ENSDART00000131875
|
atrn
|
attractin |
| chr19_+_25009921 | 0.10 |
|
|
|
| chr14_+_34626233 | 0.10 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr13_-_41821890 | 0.09 |
ENSDART00000132808
|
nanp
|
N-acetylneuraminic acid phosphatase |
| chr1_-_55765067 | 0.09 |
ENSDART00000037576
|
slc25a10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr7_-_51364562 | 0.09 |
ENSDART00000174131
|
rps4x
|
ribosomal protein S4, X-linked |
| chr8_+_27536121 | 0.09 |
ENSDART00000016696
|
rhocb
|
ras homolog family member Cb |
| chr14_+_7071129 | 0.08 |
ENSDART00000142158
|
hars
|
histidyl-tRNA synthetase |
| chr19_+_31944725 | 0.08 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr11_-_18281078 | 0.07 |
|
|
|
| chr24_+_9371496 | 0.07 |
|
|
|
| chr11_+_44805115 | 0.06 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr9_-_23177842 | 0.06 |
|
|
|
| chr25_+_35787139 | 0.06 |
ENSDART00000073402
|
si:ch211-113a14.16
|
si:ch211-113a14.16 |
| chr21_+_21575563 | 0.06 |
ENSDART00000008099
|
b9d2
|
B9 protein domain 2 |
| chr3_+_47412661 | 0.05 |
ENSDART00000155648
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr5_-_18457877 | 0.04 |
ENSDART00000142531
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr20_+_47639141 | 0.04 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr12_-_6871799 | 0.03 |
ENSDART00000171511
ENSDART00000171846 ENSDART00000166531 ENSDART00000152322 |
pcdh15b
|
protocadherin-related 15b |
| chr10_-_14582293 | 0.03 |
ENSDART00000039161
|
hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr16_+_7325399 | 0.03 |
ENSDART00000081477
|
sri
|
sorcin |
| chr17_+_53277665 | 0.02 |
ENSDART00000129236
|
wu:fe17e08
|
wu:fe17e08 |
| chr17_+_45824480 | 0.02 |
|
|
|
| chr16_+_5625788 | 0.02 |
|
|
|
| chr14_+_7596207 | 0.02 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr22_+_13862110 | 0.02 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr1_+_19376651 | 0.01 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr1_+_28860348 | 0.00 |
ENSDART00000154342
ENSDART00000088290 |
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.2 | 0.5 | GO:0002280 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.5 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.4 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.1 | 0.3 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.3 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.2 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.4 | GO:0043649 | coenzyme catabolic process(GO:0009109) dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 1.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.4 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0051427 | hormone receptor binding(GO:0051427) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |