Project
DANIO-CODE
Navigation
Downloads
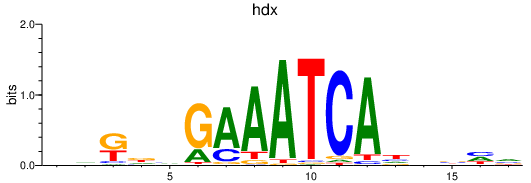
Results for hdx
Z-value: 0.78
Transcription factors associated with hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hdx
|
ENSDARG00000079382 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hdx | dr10_dc_chr14_-_5918482_5918516 | -0.70 | 2.5e-03 | Click! |
Activity profile of hdx motif
Sorted Z-values of hdx motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hdx
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_12916555 | 2.12 |
ENSDART00000140691
|
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr6_+_23476669 | 1.80 |
|
|
|
| chr21_-_2296253 | 1.73 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr5_+_52199662 | 1.71 |
|
|
|
| chr6_-_2000017 | 1.66 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr2_+_34984631 | 1.59 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr21_-_7334721 | 1.57 |
ENSDART00000136671
|
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr5_-_17502869 | 1.45 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr23_+_32174669 | 1.42 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr10_-_25608902 | 1.32 |
ENSDART00000147876
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr12_+_30673985 | 1.31 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr10_-_36849540 | 1.30 |
|
|
|
| chr16_-_26947117 | 1.29 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr5_-_63773138 | 1.27 |
ENSDART00000172321
|
brd3b
|
bromodomain containing 3b |
| chr3_-_39316317 | 1.25 |
|
|
|
| chr2_+_37312637 | 1.24 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr10_-_36849596 | 1.19 |
|
|
|
| chr2_-_54224744 | 1.14 |
|
|
|
| chr23_+_44730111 | 1.13 |
ENSDART00000177271
|
trappc1
|
trafficking protein particle complex 1 |
| chr23_+_19728953 | 1.11 |
ENSDART00000104441
|
abhd6b
|
abhydrolase domain containing 6b |
| chr7_-_13416823 | 1.09 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| chr20_-_34894930 | 1.08 |
|
|
|
| chr10_-_36849734 | 1.07 |
|
|
|
| chr11_-_24300849 | 1.07 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr14_+_31265616 | 1.06 |
|
|
|
| chr8_-_53165501 | 1.02 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr7_+_40866623 | 1.01 |
ENSDART00000052274
|
puf60b
|
poly-U binding splicing factor b |
| chr15_-_26954009 | 1.01 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr5_+_62611606 | 0.97 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr23_+_10870124 | 0.97 |
ENSDART00000035693
|
ppp4r2a
|
protein phosphatase 4, regulatory subunit 2a |
| chr12_-_26529300 | 0.96 |
ENSDART00000162163
ENSDART00000087067 |
hexdc
|
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
| chr2_+_46210971 | 0.96 |
|
|
|
| chr16_-_43491205 | 0.95 |
|
|
|
| chr7_+_15479700 | 0.95 |
|
|
|
| chr22_+_2049719 | 0.92 |
|
|
|
| chr22_+_11943005 | 0.91 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr21_+_7570595 | 0.91 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr16_-_7703633 | 0.88 |
ENSDART00000148581
|
CT574585.1
|
ENSDARG00000095852 |
| chr23_+_37476457 | 0.88 |
ENSDART00000178064
|
AL954146.3
|
ENSDARG00000108629 |
| chr11_+_8142982 | 0.87 |
ENSDART00000166379
|
samd13
|
sterile alpha motif domain containing 13 |
| chr23_-_42918086 | 0.86 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr20_-_34894607 | 0.85 |
|
|
|
| chr7_+_35163845 | 0.85 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr1_+_58608231 | 0.84 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr2_+_46210909 | 0.83 |
|
|
|
| chr7_+_38690837 | 0.82 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr15_-_6949405 | 0.78 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr16_+_42567707 | 0.78 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr14_+_30058172 | 0.77 |
ENSDART00000053925
|
mtmr7a
|
myotubularin related protein 7a |
| chr19_+_5052459 | 0.75 |
ENSDART00000003634
|
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr10_-_36849472 | 0.75 |
|
|
|
| chr23_-_42918055 | 0.73 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr8_+_36527653 | 0.72 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr5_-_13945310 | 0.71 |
ENSDART00000157675
|
kat6a
|
K(lysine) acetyltransferase 6A |
| chr2_-_40067309 | 0.71 |
|
|
|
| chr19_+_20404995 | 0.69 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr7_+_48025010 | 0.69 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr22_-_4787016 | 0.68 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr1_-_22577855 | 0.67 |
ENSDART00000175685
|
rfc1
|
replication factor C (activator 1) 1 |
| chr11_-_24300785 | 0.67 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr15_-_26953872 | 0.65 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr22_-_11024649 | 0.65 |
ENSDART00000105823
ENSDART00000159995 |
insrb
|
insulin receptor b |
| chr6_-_2000167 | 0.64 |
ENSDART00000160475
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr22_+_35156074 | 0.63 |
ENSDART00000130581
|
rnf13
|
ring finger protein 13 |
| chr17_+_6378694 | 0.63 |
ENSDART00000062952
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr3_-_59690168 | 0.62 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr1_+_58608185 | 0.62 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr22_+_35229352 | 0.62 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr1_-_20919505 | 0.61 |
|
|
|
| chr13_+_13814142 | 0.61 |
ENSDART00000142997
|
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr4_+_377994 | 0.61 |
ENSDART00000148933
|
rpl18a
|
ribosomal protein L18a |
| chr5_+_1109967 | 0.61 |
ENSDART00000046781
ENSDART00000163101 |
rnf185
|
ring finger protein 185 |
| chr2_-_37299150 | 0.61 |
ENSDART00000137598
|
nadkb
|
NAD kinase b |
| chr19_+_20405107 | 0.60 |
ENSDART00000151066
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr2_-_41035670 | 0.59 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr6_-_41023836 | 0.58 |
ENSDART00000103776
|
hdhd3
|
haloacid dehalogenase-like hydrolase domain containing 3 |
| chr3_-_33789615 | 0.58 |
|
|
|
| chr22_-_23252993 | 0.58 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr10_+_33758581 | 0.57 |
ENSDART00000141650
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr18_+_5199305 | 0.57 |
ENSDART00000020233
|
zdhhc7
|
zinc finger, DHHC-type containing 7 |
| chr5_+_56539999 | 0.56 |
ENSDART00000167660
|
pja2
|
praja ring finger 2 |
| chr18_+_9755131 | 0.55 |
|
|
|
| chr6_-_22910576 | 0.54 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr10_+_39972154 | 0.54 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr22_-_9866462 | 0.54 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr13_+_33578737 | 0.54 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr25_+_11212787 | 0.54 |
ENSDART00000159583
|
FQ312013.1
|
ENSDARG00000099473 |
| chr16_+_32128415 | 0.53 |
ENSDART00000137029
|
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr16_+_19223683 | 0.52 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr14_-_11714513 | 0.52 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr24_+_39750793 | 0.51 |
ENSDART00000133284
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr10_-_36848930 | 0.51 |
|
|
|
| KN150384v1_+_17709 | 0.51 |
|
|
|
| chr3_-_25361866 | 0.50 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr5_-_56539892 | 0.50 |
|
|
|
| chr23_-_15280634 | 0.50 |
ENSDART00000056570
|
phf20b
|
PHD finger protein 20, b |
| chr22_+_1568354 | 0.49 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr10_+_1653868 | 0.49 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr23_-_42918166 | 0.48 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr6_-_31361901 | 0.48 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr11_+_33154781 | 0.48 |
|
|
|
| chr16_-_7703754 | 0.47 |
ENSDART00000148581
|
CT574585.1
|
ENSDARG00000095852 |
| chr2_-_41035732 | 0.47 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr3_-_25361708 | 0.47 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr8_+_25988813 | 0.47 |
ENSDART00000058100
|
xpc
|
xeroderma pigmentosum, complementation group C |
| chr24_+_9740978 | 0.46 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr6_-_6138267 | 0.46 |
ENSDART00000092172
|
mtif2
|
mitochondrial translational initiation factor 2 |
| chr11_+_42310157 | 0.45 |
ENSDART00000085868
|
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr6_-_41023783 | 0.44 |
ENSDART00000103776
|
hdhd3
|
haloacid dehalogenase-like hydrolase domain containing 3 |
| chr13_+_46429054 | 0.44 |
ENSDART00000160401
|
tmem63ba
|
transmembrane protein 63Ba |
| chr5_+_13146987 | 0.43 |
ENSDART00000139199
|
h2afva
|
H2A histone family, member Va |
| chr1_+_43822319 | 0.42 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr3_-_52837780 | 0.42 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr9_-_12678315 | 0.42 |
|
|
|
| chr22_-_4786947 | 0.42 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr14_-_30150071 | 0.41 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr22_+_35229139 | 0.39 |
ENSDART00000061315
ENSDART00000146430 |
tsc22d2
|
TSC22 domain family 2 |
| chr23_-_465425 | 0.39 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr13_+_9510368 | 0.39 |
ENSDART00000140504
|
prdx3
|
peroxiredoxin 3 |
| chr7_-_13416289 | 0.39 |
|
|
|
| chr7_+_17701669 | 0.37 |
ENSDART00000113120
|
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr12_+_26785950 | 0.37 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr12_+_3834709 | 0.37 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr15_-_26954063 | 0.37 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr8_-_17130906 | 0.36 |
|
|
|
| chr25_-_23485520 | 0.36 |
ENSDART00000149107
|
nap1l4a
|
nucleosome assembly protein 1-like 4a |
| chr17_-_970957 | 0.36 |
ENSDART00000177536
|
dnajc17
|
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr6_+_8078889 | 0.35 |
ENSDART00000134245
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr5_-_6182820 | 0.35 |
ENSDART00000168698
|
ostf1
|
osteoclast stimulating factor 1 |
| chr20_+_50249994 | 0.34 |
ENSDART00000047212
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr6_+_40566498 | 0.34 |
ENSDART00000154766
|
si:ch73-15b2.5
|
si:ch73-15b2.5 |
| chr24_+_39769129 | 0.33 |
ENSDART00000066500
ENSDART00000145075 |
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr5_+_25080638 | 0.32 |
ENSDART00000134242
|
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr5_-_63773215 | 0.32 |
ENSDART00000168030
|
brd3b
|
bromodomain containing 3b |
| chr9_+_12917201 | 0.32 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr5_+_61278798 | 0.31 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr5_+_36066523 | 0.31 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr13_+_18377514 | 0.30 |
|
|
|
| chr10_-_36849108 | 0.30 |
|
|
|
| chr9_-_2756749 | 0.30 |
|
|
|
| chr6_+_54568649 | 0.30 |
ENSDART00000157142
|
tead3b
|
TEA domain family member 3 b |
| chr2_+_31823314 | 0.30 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr16_+_42567668 | 0.29 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr10_-_44441197 | 0.29 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr17_+_6378932 | 0.28 |
ENSDART00000062952
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr7_+_15479520 | 0.28 |
|
|
|
| chr24_-_36248243 | 0.28 |
ENSDART00000065336
|
tmem14cb
|
transmembrane protein 14Cb |
| chr3_+_33788977 | 0.27 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr12_+_30119447 | 0.27 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr9_-_32532616 | 0.26 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr6_+_54568388 | 0.26 |
ENSDART00000093199
|
tead3b
|
TEA domain family member 3 b |
| chr23_-_39879096 | 0.25 |
ENSDART00000159519
|
ENSDARG00000102199
|
ENSDARG00000102199 |
| chr16_+_19223553 | 0.25 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr24_+_33478309 | 0.25 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr9_+_26029780 | 0.24 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr7_+_34216448 | 0.24 |
ENSDART00000109872
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr9_+_56868126 | 0.24 |
ENSDART00000160107
|
CU639413.1
|
ENSDARG00000100275 |
| chr6_-_40165125 | 0.23 |
ENSDART00000153956
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr22_+_18904372 | 0.23 |
ENSDART00000131131
|
bsg
|
basigin |
| chr25_+_3619832 | 0.23 |
|
|
|
| chr11_+_42184049 | 0.22 |
ENSDART00000056048
|
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr5_-_17502894 | 0.21 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr22_+_35156212 | 0.21 |
ENSDART00000131952
ENSDART00000003303 |
rnf13
|
ring finger protein 13 |
| chr6_+_6674857 | 0.20 |
ENSDART00000151343
|
ENSDARG00000077040
|
ENSDARG00000077040 |
| chr18_-_22105000 | 0.20 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr16_+_23580507 | 0.19 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr5_-_7591027 | 0.19 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr7_+_19779221 | 0.19 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr24_-_24999348 | 0.18 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr16_+_27449058 | 0.18 |
ENSDART00000132329
|
stx17
|
syntaxin 17 |
| chr16_+_51290761 | 0.18 |
ENSDART00000058290
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr19_-_24971920 | 0.18 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr25_+_15901561 | 0.17 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr8_+_36527535 | 0.16 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr9_-_32489944 | 0.16 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr10_+_8142976 | 0.16 |
ENSDART00000123447
|
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr3_-_25361754 | 0.16 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr1_-_28146457 | 0.16 |
ENSDART00000110270
ENSDART00000170797 |
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr24_+_39768911 | 0.16 |
ENSDART00000066500
ENSDART00000145075 |
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr25_+_10714737 | 0.15 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr5_+_26820868 | 0.13 |
ENSDART00000098590
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr5_-_23179648 | 0.13 |
|
|
|
| chr25_+_35784138 | 0.12 |
ENSDART00000152761
|
CR354435.7
|
ENSDARG00000094744 |
| chr1_-_22577723 | 0.12 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr4_-_72627654 | 0.12 |
ENSDART00000112820
|
zgc:165515
|
zgc:165515 |
| chr18_+_5199209 | 0.11 |
ENSDART00000020233
|
zdhhc7
|
zinc finger, DHHC-type containing 7 |
| chr10_+_9512758 | 0.11 |
ENSDART00000141869
|
rbm18
|
RNA binding motif protein 18 |
| chr19_+_19164500 | 0.11 |
ENSDART00000165728
|
vps52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr11_+_42310184 | 0.10 |
ENSDART00000085868
|
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr7_-_48123296 | 0.10 |
ENSDART00000174381
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr11_+_30049041 | 0.09 |
|
|
|
| chr14_+_26421549 | 0.09 |
ENSDART00000172927
|
eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr16_+_39292206 | 0.08 |
ENSDART00000127319
ENSDART00000102510 |
zgc:77056
|
zgc:77056 |
| chr25_+_17765920 | 0.08 |
ENSDART00000151853
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr20_+_21578759 | 0.08 |
|
|
|
| chr8_-_16702377 | 0.07 |
ENSDART00000100706
|
ndufaf2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr5_+_71409912 | 0.07 |
ENSDART00000165835
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr3_+_26092485 | 0.07 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr24_-_17255674 | 0.06 |
ENSDART00000134947
ENSDART00000122757 |
cul1b
|
cullin 1b |
| chr4_+_26047568 | 0.06 |
ENSDART00000126474
|
si:ch211-265o23.1
|
si:ch211-265o23.1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 0.7 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.3 | 0.8 | GO:0070655 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.3 | 1.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 1.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.9 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 1.3 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 2.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0032675 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) |
| 0.1 | 2.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.5 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.1 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 2.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.5 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) regulation of steroid metabolic process(GO:0019218) |
| 0.1 | 1.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.7 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 1.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.3 | GO:0070265 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.4 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.5 | GO:0071005 | U4 snRNP(GO:0005687) U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 1.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.0 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.5 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 0.9 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 1.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 0.7 | GO:0019705 | protein-cysteine S-myristoyltransferase activity(GO:0019705) |
| 0.2 | 2.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 2.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.6 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 2.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |