Project
DANIO-CODE
Navigation
Downloads
Results for her6+her8.2+her9
Z-value: 0.32
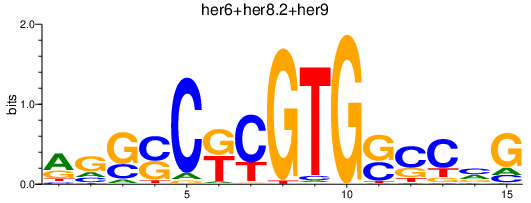
Transcription factors associated with her6+her8.2+her9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
her6
|
ENSDARG00000006514 | hairy-related 6 |
|
her9
|
ENSDARG00000056438 | hairy-related 9 |
|
her8.2
|
ENSDARG00000069675 | hairy-related 8.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| her9 | dr10_dc_chr23_-_23474703_23474754 | 0.25 | 3.4e-01 | Click! |
| her6 | dr10_dc_chr6_-_36574848_36574899 | 0.08 | 7.6e-01 | Click! |
| her8.2 | dr10_dc_chr15_+_7179382_7179411 | 0.07 | 7.8e-01 | Click! |
Activity profile of her6+her8.2+her9 motif
Sorted Z-values of her6+her8.2+her9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of her6+her8.2+her9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_29846530 | 0.49 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_+_24770873 | 0.44 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr23_+_45020761 | 0.38 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr4_+_830826 | 0.36 |
|
|
|
| chr5_+_34798432 | 0.35 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr15_-_47234665 | 0.29 |
ENSDART00000124474
|
CU633855.1
|
ENSDARG00000089563 |
| chr21_-_2116499 | 0.28 |
ENSDART00000167307
|
AL627305.3
|
ENSDARG00000102019 |
| chr24_+_26287471 | 0.27 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr23_-_19053587 | 0.25 |
|
|
|
| chr23_-_6831711 | 0.24 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr16_+_11138879 | 0.21 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr5_+_1699963 | 0.21 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr11_-_41219340 | 0.20 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr16_+_11138924 | 0.20 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr17_+_612173 | 0.19 |
|
|
|
| chr3_-_58764372 | 0.19 |
ENSDART00000108869
|
luc7l3
|
LUC7-like 3 pre-mRNA splicing factor |
| chr25_-_8436134 | 0.18 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr7_-_6302741 | 0.18 |
ENSDART00000173180
|
hist1h4l
|
histone 1, H4, like |
| chr15_+_14925920 | 0.17 |
|
|
|
| chr9_+_9133904 | 0.17 |
ENSDART00000165932
|
sik1
|
salt-inducible kinase 1 |
| chr3_-_29739166 | 0.14 |
ENSDART00000147048
|
rpl3
|
ribosomal protein L3 |
| chr5_-_1698166 | 0.14 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr18_+_16136765 | 0.14 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr24_-_33377293 | 0.13 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr21_-_44707514 | 0.12 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr19_+_48555386 | 0.12 |
|
|
|
| chr15_+_14925529 | 0.12 |
ENSDART00000166250
|
CR936977.2
|
ENSDARG00000103638 |
| chr19_+_48555339 | 0.11 |
|
|
|
| chr21_-_44708217 | 0.10 |
|
|
|
| chr7_-_68961464 | 0.10 |
ENSDART00000168311
|
usp10
|
ubiquitin specific peptidase 10 |
| chr11_-_41219410 | 0.09 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr5_-_54026450 | 0.09 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr23_+_22649686 | 0.08 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr9_-_43736549 | 0.08 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr1_+_43689095 | 0.07 |
ENSDART00000100321
|
timm10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr25_-_21058792 | 0.07 |
ENSDART00000156257
|
wnk1a
|
WNK lysine deficient protein kinase 1a |
| chr23_+_22649652 | 0.07 |
ENSDART00000146709
|
CR388207.1
|
ENSDARG00000095377 |
| chr15_-_1800139 | 0.06 |
ENSDART00000093074
|
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr2_+_58654570 | 0.06 |
ENSDART00000164102
ENSDART00000158777 |
cirbpa
|
cold inducible RNA binding protein a |
| chr12_+_19240407 | 0.06 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr5_+_33011105 | 0.05 |
|
|
|
| chr1_-_51826194 | 0.05 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr20_-_54942216 | 0.05 |
|
|
|
| chr23_+_36494295 | 0.05 |
ENSDART00000042701
|
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr25_+_27300835 | 0.05 |
ENSDART00000103519
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr13_-_44885345 | 0.04 |
ENSDART00000074787
ENSDART00000125633 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr3_-_29846789 | 0.04 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr2_-_40067342 | 0.04 |
|
|
|
| chr9_+_492585 | 0.04 |
ENSDART00000112635
|
ENSDARG00000078172
|
ENSDARG00000078172 |
| chr1_-_51826431 | 0.03 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr14_-_28261949 | 0.03 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr15_-_35802739 | 0.02 |
ENSDART00000174714
|
CR848683.2
|
ENSDARG00000106192 |
| chr9_-_43087163 | 0.02 |
|
|
|
| chr10_-_7862803 | 0.02 |
ENSDART00000059021
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr23_+_36494057 | 0.02 |
ENSDART00000042701
|
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr3_+_14492079 | 0.02 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr13_-_24695644 | 0.01 |
ENSDART00000087786
|
slka
|
STE20-like kinase a |
| chr24_-_26339981 | 0.01 |
ENSDART00000135496
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr25_-_29020637 | 0.01 |
ENSDART00000124645
|
ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr20_-_6153133 | 0.00 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr5_-_32290543 | 0.00 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr14_-_24113237 | 0.00 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.0 | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.0 | 0.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |