Project
DANIO-CODE
Navigation
Downloads
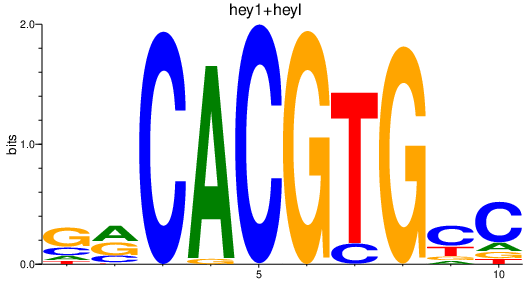
Results for hey1+heyl_hey2
Z-value: 0.55
Transcription factors associated with hey1+heyl_hey2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
heyl
|
ENSDARG00000055798 | hes related family bHLH transcription factor with YRPW motif like |
|
hey1
|
ENSDARG00000070538 | hes-related family bHLH transcription factor with YRPW motif 1 |
|
hey2
|
ENSDARG00000013441 | hes-related family bHLH transcription factor with YRPW motif 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hey1 | dr10_dc_chr19_-_32214978_32214990 | 0.31 | 2.4e-01 | Click! |
| hey2 | dr10_dc_chr20_-_39693535_39693592 | 0.23 | 4.0e-01 | Click! |
Activity profile of hey1+heyl_hey2 motif
Sorted Z-values of hey1+heyl_hey2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hey1+heyl_hey2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_14531021 | 0.83 |
ENSDART00000170954
|
si:dkey-102m7.3
|
si:dkey-102m7.3 |
| chr19_-_24971633 | 0.68 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr25_-_34608926 | 0.62 |
ENSDART00000130395
|
hist1h4l
|
histone 1, H4, like |
| chr25_-_35817806 | 0.55 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr7_+_24770873 | 0.52 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr20_-_25726868 | 0.49 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr2_-_24661477 | 0.49 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr7_+_58718614 | 0.48 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr8_+_17148864 | 0.48 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr12_-_7572970 | 0.48 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr10_+_4962403 | 0.46 |
ENSDART00000134679
|
CU074419.2
|
ENSDARG00000093688 |
| chr6_-_47842137 | 0.46 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr19_+_7233537 | 0.44 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr10_-_8029671 | 0.44 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr22_+_16471319 | 0.44 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr10_-_44441481 | 0.44 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr20_+_32503118 | 0.44 |
ENSDART00000018640
|
snx3
|
sorting nexin 3 |
| chr2_+_25363717 | 0.44 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr20_-_23184142 | 0.43 |
ENSDART00000176282
|
CU693368.2
|
ENSDARG00000108718 |
| chr11_-_16840339 | 0.43 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr11_+_3940085 | 0.42 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr1_-_674449 | 0.42 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr19_+_42899678 | 0.41 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr15_+_1187249 | 0.41 |
ENSDART00000152638
ENSDART00000152466 |
mlf1
|
myeloid leukemia factor 1 |
| chr16_-_45103102 | 0.41 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_-_791149 | 0.38 |
ENSDART00000151782
ENSDART00000037515 |
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr10_-_22065636 | 0.38 |
|
|
|
| chr16_+_43468861 | 0.38 |
|
|
|
| chr24_+_32754013 | 0.37 |
ENSDART00000156638
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr4_-_21931540 | 0.37 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr11_+_30416953 | 0.36 |
|
|
|
| chr13_+_46514162 | 0.36 |
ENSDART00000159260
|
CU695232.1
|
ENSDARG00000098286 |
| chr7_-_73530653 | 0.35 |
ENSDART00000166633
ENSDART00000009888 ENSDART00000171254 |
casq1b
|
calsequestrin 1b |
| chr19_+_43524098 | 0.35 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr13_+_31271568 | 0.35 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr9_-_41044622 | 0.34 |
ENSDART00000143384
|
si:dkey-95p16.1
|
si:dkey-95p16.1 |
| chr11_+_44922098 | 0.34 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr13_-_18506153 | 0.33 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr12_+_24221087 | 0.33 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr24_-_12794564 | 0.33 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr8_+_23192085 | 0.33 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr20_+_45989418 | 0.32 |
ENSDART00000131169
|
bmp2b
|
bone morphogenetic protein 2b |
| chr22_-_120677 | 0.31 |
|
|
|
| chr19_+_7234029 | 0.31 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr21_-_20728623 | 0.30 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr6_-_28231995 | 0.29 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr3_+_1455113 | 0.29 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr21_-_20674965 | 0.28 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr17_+_27383737 | 0.28 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr7_-_6205157 | 0.28 |
ENSDART00000172884
|
hist1h4l
|
histone 1, H4, like |
| chr24_-_26339981 | 0.28 |
ENSDART00000135496
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr5_+_4033390 | 0.28 |
ENSDART00000149185
|
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr20_+_54464026 | 0.27 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr24_-_12794672 | 0.27 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_-_60152594 | 0.27 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr8_+_166461 | 0.26 |
|
|
|
| chr5_-_41039135 | 0.26 |
ENSDART00000083644
|
epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr9_+_24255064 | 0.26 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr24_-_33869817 | 0.26 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr6_-_60152693 | 0.25 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr3_-_10455344 | 0.25 |
ENSDART00000111833
|
pctp
|
phosphatidylcholine transfer protein |
| chr3_+_54327353 | 0.25 |
ENSDART00000127487
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr19_+_34145030 | 0.25 |
ENSDART00000151521
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_-_19114558 | 0.25 |
ENSDART00000177621
|
dennd4a
|
DENN/MADD domain containing 4A |
| chr19_+_43523303 | 0.24 |
ENSDART00000167847
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr7_+_44373815 | 0.24 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr4_+_11054808 | 0.24 |
ENSDART00000140362
|
ccdc59
|
coiled-coil domain containing 59 |
| chr20_-_48677794 | 0.24 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr9_-_785511 | 0.23 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| KN150670v1_-_72156 | 0.23 |
|
|
|
| chr3_+_1455242 | 0.23 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr8_+_23716843 | 0.23 |
ENSDART00000136547
|
rpl10a
|
ribosomal protein L10a |
| chr3_+_32394653 | 0.23 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr2_-_9691594 | 0.23 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr5_+_23625065 | 0.23 |
ENSDART00000029719
|
si:ch211-114c12.2
|
si:ch211-114c12.2 |
| chr7_-_29811734 | 0.23 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr23_-_32043229 | 0.23 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr1_+_16683931 | 0.22 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr18_+_9524331 | 0.22 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| KN150487v1_+_15409 | 0.22 |
ENSDART00000166996
|
CABZ01074304.1
|
ENSDARG00000100224 |
| chr14_-_31676235 | 0.22 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr8_+_20406797 | 0.22 |
ENSDART00000016422
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr2_-_42525768 | 0.22 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr6_+_42821679 | 0.22 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr18_-_17031173 | 0.22 |
ENSDART00000129146
|
tbc1d15
|
TBC1 domain family, member 15 |
| chr2_+_5297505 | 0.22 |
ENSDART00000108990
|
pex5lb
|
peroxisomal biogenesis factor 5-like b |
| chr9_+_54692834 | 0.22 |
|
|
|
| chr12_-_22438379 | 0.22 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr24_-_8592157 | 0.21 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr11_+_7139675 | 0.21 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr8_-_17148743 | 0.21 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr23_+_35964754 | 0.21 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr5_+_22173976 | 0.21 |
ENSDART00000142112
|
vma21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr18_-_21229799 | 0.21 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr24_+_12845250 | 0.21 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr21_-_776741 | 0.20 |
|
|
|
| chr16_+_7213011 | 0.20 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr19_-_9192541 | 0.20 |
|
|
|
| chr1_-_22144014 | 0.20 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr25_+_36872560 | 0.20 |
ENSDART00000163178
|
slc10a3
|
solute carrier family 10, member 3 |
| chr7_-_28376722 | 0.20 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr4_-_16835283 | 0.20 |
ENSDART00000165289
|
gys2
|
glycogen synthase 2 |
| chr12_+_30471781 | 0.20 |
ENSDART00000126984
|
nrap
|
nebulin-related anchoring protein |
| chr16_-_15496868 | 0.19 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr10_+_29351676 | 0.19 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr5_+_2384929 | 0.19 |
|
|
|
| chr2_+_50126676 | 0.19 |
ENSDART00000145483
|
rpl37
|
ribosomal protein L37 |
| chr12_+_19262722 | 0.19 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr3_+_28871566 | 0.19 |
|
|
|
| chr1_-_54087595 | 0.19 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr18_+_14725148 | 0.18 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr7_-_40713381 | 0.18 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr18_+_9524247 | 0.18 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr4_-_13922285 | 0.18 |
ENSDART00000080334
|
yaf2
|
YY1 associated factor 2 |
| chr18_-_26876598 | 0.18 |
|
|
|
| chr13_-_40190349 | 0.18 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr7_+_67244332 | 0.18 |
ENSDART00000170322
|
rpl13
|
ribosomal protein L13 |
| chr15_-_2689005 | 0.18 |
ENSDART00000063325
|
cldnf
|
claudin f |
| chr15_+_16961487 | 0.18 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr5_-_9162684 | 0.17 |
|
|
|
| chr24_+_23599223 | 0.17 |
|
|
|
| chr10_+_5422223 | 0.17 |
ENSDART00000063093
|
auh
|
AU RNA binding protein/enoyl-CoA hydratase |
| chr1_-_17073539 | 0.17 |
|
|
|
| chr11_+_39768718 | 0.17 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr23_+_7614867 | 0.17 |
ENSDART00000011554
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr6_-_12883682 | 0.17 |
ENSDART00000165375
|
zgc:194469
|
zgc:194469 |
| chr10_-_11053655 | 0.17 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr25_-_20994084 | 0.17 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| KN150680v1_+_10854 | 0.17 |
|
|
|
| chr16_+_53632229 | 0.17 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr20_+_1887099 | 0.17 |
|
|
|
| chr24_-_12794057 | 0.17 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr12_-_9479063 | 0.17 |
ENSDART00000169727
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr18_+_20001243 | 0.17 |
ENSDART00000090310
|
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr15_+_19902697 | 0.17 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr20_-_128304 | 0.17 |
ENSDART00000149581
|
ypel5
|
yippee-like 5 |
| chr18_+_44802349 | 0.17 |
ENSDART00000139526
|
fam118b
|
family with sequence similarity 118, member B |
| chr2_-_58737903 | 0.17 |
|
|
|
| chr6_+_60190849 | 0.16 |
|
|
|
| chr1_+_30142256 | 0.16 |
ENSDART00000022841
|
metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr22_+_17581859 | 0.16 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr8_-_387126 | 0.16 |
|
|
|
| chr21_+_713673 | 0.16 |
|
|
|
| chr16_-_24697750 | 0.16 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr18_-_5260386 | 0.16 |
ENSDART00000027115
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr1_-_50215233 | 0.16 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr22_+_18228143 | 0.16 |
ENSDART00000141535
|
BX664610.1
|
ENSDARG00000095557 |
| chr18_+_9524184 | 0.16 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr19_-_24971920 | 0.16 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr18_+_5260471 | 0.16 |
ENSDART00000150992
|
wdr59
|
WD repeat domain 59 |
| chr20_-_5324678 | 0.16 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr17_+_19479310 | 0.16 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr6_+_60152913 | 0.15 |
|
|
|
| chr25_-_35817715 | 0.15 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr12_-_34621359 | 0.15 |
|
|
|
| chr8_-_31597252 | 0.15 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr19_+_2754903 | 0.15 |
ENSDART00000160533
|
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr9_+_56881036 | 0.15 |
|
|
|
| chr10_+_22942760 | 0.15 |
ENSDART00000134790
|
med11
|
mediator complex subunit 11 |
| chr6_-_57479360 | 0.15 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr20_+_27341026 | 0.15 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr19_+_34582100 | 0.15 |
ENSDART00000135592
|
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr25_+_222244 | 0.15 |
ENSDART00000155344
|
ENSDARG00000073905
|
ENSDARG00000073905 |
| chr11_-_42826276 | 0.15 |
|
|
|
| chr4_+_830826 | 0.15 |
|
|
|
| chr24_+_42133471 | 0.15 |
ENSDART00000170514
|
top1mt
|
topoisomerase (DNA) I, mitochondrial |
| chr12_+_27194414 | 0.15 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr16_+_24045707 | 0.15 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr8_-_16577737 | 0.15 |
ENSDART00000139038
|
CU467861.1
|
ENSDARG00000092337 |
| chr20_-_22478716 | 0.15 |
ENSDART00000110967
ENSDART00000011135 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr7_+_5846555 | 0.15 |
ENSDART00000083397
|
hist1h4l
|
histone 1, H4, like |
| chr1_+_157674 | 0.14 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr5_+_68410884 | 0.14 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr16_+_22950567 | 0.14 |
ENSDART00000143957
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr7_-_71092394 | 0.14 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr1_+_30840656 | 0.14 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr3_-_34208423 | 0.14 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr10_+_5267746 | 0.14 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr24_-_8592102 | 0.14 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr13_+_24531753 | 0.14 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr17_+_53068731 | 0.14 |
ENSDART00000156774
|
dph6
|
diphthamine biosynthesis 6 |
| chr13_-_25068717 | 0.14 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr20_-_158899 | 0.14 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr3_+_53518347 | 0.14 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr22_-_520129 | 0.14 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr25_+_18866796 | 0.14 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| KN150670v1_-_61400 | 0.14 |
|
|
|
| chr23_+_2482102 | 0.14 |
ENSDART00000126038
|
tcp1
|
t-complex 1 |
| chr24_-_42120772 | 0.14 |
ENSDART00000166413
ENSDART00000166414 |
ssr1
|
signal sequence receptor, alpha |
| chr9_-_9106512 | 0.14 |
|
|
|
| chr12_-_47699958 | 0.13 |
ENSDART00000171932
ENSDART00000168165 ENSDART00000161985 |
HHEX
|
hematopoietically expressed homeobox |
| chr24_-_3751435 | 0.13 |
ENSDART00000139596
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr19_+_33235087 | 0.13 |
ENSDART00000052097
ENSDART00000052096 |
hrsp12
|
heat-responsive protein 12 |
| chr16_+_7213161 | 0.13 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr17_+_48734060 | 0.13 |
ENSDART00000178697
|
daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr20_-_9212177 | 0.13 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr7_-_32562435 | 0.13 |
ENSDART00000099872
ENSDART00000099871 ENSDART00000147554 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr22_-_120910 | 0.13 |
|
|
|
| chr12_-_4497094 | 0.13 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr18_+_41570495 | 0.13 |
ENSDART00000169621
ENSDART00000162052 |
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.4 | GO:0061130 | branching involved in pancreas morphogenesis(GO:0061114) pancreatic bud formation(GO:0061130) pancreas field specification(GO:0061131) |
| 0.1 | 0.4 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 0.6 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.4 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.3 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.2 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.3 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.2 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.2 | GO:0046379 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.0 | 0.2 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.4 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.1 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.1 | GO:0006210 | thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0019184 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 1.2 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.5 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.2 | 0.5 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.4 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.3 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |