Project
DANIO-CODE
Navigation
Downloads
Results for hic1l+hic2
Z-value: 0.25
Transcription factors associated with hic1l+hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1l
|
ENSDARG00000045660 | hypermethylated in cancer 1 like |
|
hic2
|
ENSDARG00000100497 | hypermethylated in cancer 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic2 | dr10_dc_chr10_-_3176372_3176450 | 0.47 | 6.8e-02 | Click! |
| hic1l | dr10_dc_chr25_-_25480657_25480661 | 0.26 | 3.2e-01 | Click! |
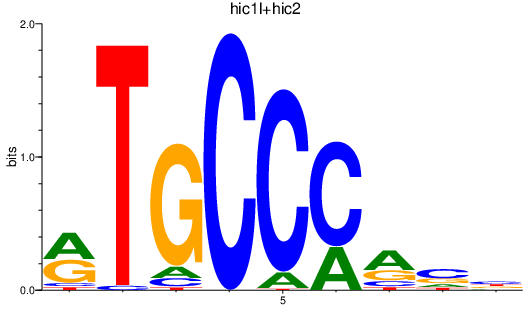
Activity profile of hic1l+hic2 motif
Sorted Z-values of hic1l+hic2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1l+hic2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_25592743 | 0.74 |
ENSDART00000110638
ENSDART00000172092 |
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr2_+_25281122 | 0.31 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr2_+_24982557 | 0.25 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr2_+_24982611 | 0.21 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr8_-_49227232 | 0.21 |
|
|
|
| chr2_+_24982203 | 0.21 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr8_-_49227286 | 0.20 |
|
|
|
| chr23_-_6831711 | 0.19 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr5_-_24013767 | 0.18 |
ENSDART00000003957
|
tia1l
|
cytotoxic granule-associated RNA binding protein 1, like |
| chr21_+_33216466 | 0.18 |
ENSDART00000163808
|
ndst1b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1b |
| chr2_+_24982471 | 0.18 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr5_-_54097015 | 0.18 |
|
|
|
| chr9_+_33409580 | 0.18 |
ENSDART00000100893
|
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr7_-_73620443 | 0.17 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr15_-_7340623 | 0.17 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
solute carrier family 7 member 1 |
| chr17_+_51851738 | 0.16 |
ENSDART00000053422
|
ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr3_+_14362849 | 0.16 |
ENSDART00000161403
|
rab3db
|
RAB3D, member RAS oncogene family, b |
| chr1_+_29919378 | 0.16 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr2_-_30929469 | 0.16 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr1_-_12575687 | 0.15 |
|
|
|
| chr7_-_33414221 | 0.13 |
|
|
|
| chr13_+_15669704 | 0.13 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr2_+_24982268 | 0.12 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr22_-_20236913 | 0.12 |
ENSDART00000134485
|
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr21_+_33216833 | 0.12 |
|
|
|
| chr6_-_19813235 | 0.12 |
ENSDART00000151152
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr7_+_22540012 | 0.12 |
|
|
|
| chr21_+_33216675 | 0.11 |
ENSDART00000163808
|
ndst1b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1b |
| chr13_+_15669924 | 0.11 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr21_+_33216241 | 0.11 |
ENSDART00000169972
|
ndst1b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1b |
| chr7_-_21620707 | 0.11 |
ENSDART00000019699
|
mettl3
|
methyltransferase like 3 |
| chr13_-_6150836 | 0.11 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr19_-_2148053 | 0.10 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr7_-_21620664 | 0.10 |
ENSDART00000019699
|
mettl3
|
methyltransferase like 3 |
| chr14_-_25636795 | 0.10 |
|
|
|
| chr8_-_1154004 | 0.09 |
ENSDART00000149969
ENSDART00000016800 |
znf367
|
zinc finger protein 367 |
| chr12_+_33793468 | 0.09 |
ENSDART00000130853
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr4_+_17666992 | 0.09 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr22_-_21873054 | 0.09 |
|
|
|
| chr20_-_54646114 | 0.08 |
ENSDART00000038745
|
yy1b
|
YY1 transcription factor b |
| chr19_+_21124187 | 0.08 |
ENSDART00000129730
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr7_-_46509861 | 0.08 |
ENSDART00000166565
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr12_+_33793663 | 0.08 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr8_+_26838596 | 0.07 |
|
|
|
| chr24_-_30924897 | 0.07 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr23_-_20199769 | 0.07 |
ENSDART00000147725
|
tktb
|
transketolase b |
| chr7_-_65012626 | 0.07 |
|
|
|
| chr21_-_38670059 | 0.07 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr22_-_5791916 | 0.07 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr3_-_30955799 | 0.06 |
ENSDART00000103421
|
zgc:153292
|
zgc:153292 |
| chr18_-_16134258 | 0.06 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr5_+_25133592 | 0.06 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr16_-_53978295 | 0.06 |
ENSDART00000158047
|
ZNRF2
|
zinc and ring finger 2 |
| chr18_-_35590515 | 0.06 |
ENSDART00000150351
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr13_-_33077267 | 0.06 |
ENSDART00000146138
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr12_+_27445044 | 0.06 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr7_+_20829815 | 0.06 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr11_-_11591523 | 0.06 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr5_-_34664282 | 0.06 |
ENSDART00000114981
ENSDART00000051279 |
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr6_-_19556328 | 0.06 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr18_-_16134320 | 0.05 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr9_+_29832294 | 0.05 |
ENSDART00000023210
|
trim13
|
tripartite motif containing 13 |
| chr9_-_22371512 | 0.05 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr1_+_29919729 | 0.05 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr23_-_20199837 | 0.05 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr11_-_41751050 | 0.04 |
ENSDART00000173086
|
zgc:175088
|
zgc:175088 |
| chr6_+_20595096 | 0.04 |
|
|
|
| chr25_-_13562597 | 0.04 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr9_-_29832812 | 0.04 |
ENSDART00000101177
|
spryd7b
|
SPRY domain containing 7b |
| chr17_+_13078286 | 0.04 |
ENSDART00000149779
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr2_-_32404140 | 0.04 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr6_-_19812488 | 0.04 |
ENSDART00000151179
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr7_+_29681510 | 0.04 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr7_+_29681196 | 0.04 |
ENSDART00000039657
|
tpma
|
alpha-tropomyosin |
| chr5_-_21520111 | 0.04 |
|
|
|
| chr17_-_24872401 | 0.03 |
ENSDART00000135569
|
gale
|
UDP-galactose-4-epimerase |
| chr24_-_39970897 | 0.03 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr2_+_11423028 | 0.03 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr2_-_30929321 | 0.03 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr25_+_11013469 | 0.03 |
ENSDART00000112789
|
AKAP13
|
A-kinase anchoring protein 13 |
| chr15_-_29415309 | 0.03 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr4_-_9821525 | 0.03 |
ENSDART00000131357
ENSDART00000138321 |
parp12b
|
poly (ADP-ribose) polymerase family, member 12b |
| chr22_-_21872864 | 0.03 |
ENSDART00000158501
|
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr13_+_290930 | 0.03 |
ENSDART00000017854
|
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr12_+_31614163 | 0.03 |
ENSDART00000152973
|
RNF157
|
ring finger protein 157 |
| chr25_-_19476079 | 0.02 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr25_-_13562558 | 0.02 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr25_-_13562693 | 0.02 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr6_-_27118157 | 0.02 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr22_+_3989571 | 0.02 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr9_-_22402341 | 0.02 |
ENSDART00000110656
|
crygm2d20
|
crystallin, gamma M2d20 |
| chr11_+_24110796 | 0.02 |
ENSDART00000089747
|
nfs1
|
NFS1 cysteine desulfurase |
| chr2_+_25281087 | 0.02 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr13_+_18377514 | 0.02 |
|
|
|
| chr17_+_8831186 | 0.02 |
ENSDART00000109573
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr19_+_48166365 | 0.02 |
|
|
|
| chr22_-_5791865 | 0.02 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr21_-_14950265 | 0.02 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr17_+_32408151 | 0.02 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr20_+_31372117 | 0.02 |
|
|
|
| chr20_-_28865097 | 0.01 |
ENSDART00000114611
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr17_-_43409222 | 0.01 |
ENSDART00000156033
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr2_-_44402452 | 0.01 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr14_+_5852428 | 0.01 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr19_-_2147899 | 0.01 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr3_-_11861502 | 0.01 |
ENSDART00000018159
|
ENSDARG00000011609
|
ENSDARG00000011609 |
| chr5_-_25133229 | 0.01 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr17_+_52736844 | 0.01 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr8_+_8934762 | 0.01 |
ENSDART00000133137
|
bcap31
|
B-cell receptor-associated protein 31 |
| chr25_+_16849382 | 0.01 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr1_+_29919605 | 0.00 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr20_-_54645903 | 0.00 |
ENSDART00000038745
|
yy1b
|
YY1 transcription factor b |
| chr3_-_11861438 | 0.00 |
ENSDART00000018159
|
ENSDARG00000011609
|
ENSDARG00000011609 |
| chr15_-_20795531 | 0.00 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr22_+_9493668 | 0.00 |
ENSDART00000143953
|
strip1
|
striatin interacting protein 1 |
| chr14_-_20766762 | 0.00 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr4_-_14916491 | 0.00 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060853 | forebrain radial glial cell differentiation(GO:0021861) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.0 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |