Project
DANIO-CODE
Navigation
Downloads
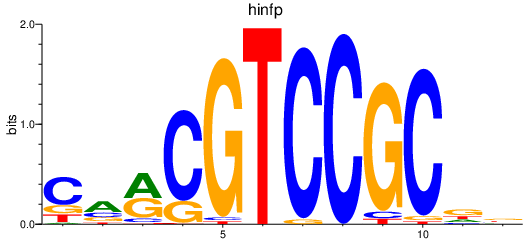
Results for hinfp
Z-value: 0.85
Transcription factors associated with hinfp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hinfp
|
ENSDARG00000004851 | histone H4 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hinfp | dr10_dc_chr5_+_29996321_29996501 | 0.45 | 7.8e-02 | Click! |
Activity profile of hinfp motif
Sorted Z-values of hinfp motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hinfp
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_6012910 | 2.44 |
ENSDART00000161001
|
gtpbp3
|
GTP binding protein 3 |
| chr9_+_8429700 | 1.65 |
ENSDART00000144373
|
zgc:153499
|
zgc:153499 |
| chr23_+_46000362 | 1.62 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr5_-_47323502 | 1.57 |
ENSDART00000175026
|
ccnh
|
cyclin H |
| chr19_+_8693855 | 1.50 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr10_-_21404605 | 1.41 |
ENSDART00000125167
|
avd
|
avidin |
| chr19_+_32178335 | 1.27 |
|
|
|
| chr19_+_8693695 | 1.21 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr6_-_1315025 | 1.14 |
|
|
|
| KN149861v1_-_5655 | 1.13 |
|
|
|
| chr20_+_35305119 | 1.10 |
ENSDART00000045135
|
fbxo16
|
F-box protein 16 |
| chr25_+_4414838 | 1.10 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr5_-_47323448 | 1.09 |
ENSDART00000175026
|
ccnh
|
cyclin H |
| chr3_+_17783319 | 1.01 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr5_+_47211204 | 0.97 |
ENSDART00000139824
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| KN149861v1_-_6089 | 0.97 |
|
|
|
| chr20_-_2708839 | 0.85 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr20_-_2708931 | 0.83 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr11_-_25615491 | 0.82 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr16_-_27629513 | 0.81 |
ENSDART00000078297
|
zgc:153215
|
zgc:153215 |
| chr8_+_11649521 | 0.80 |
ENSDART00000167298
|
atp2a2a
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2a |
| chr23_+_43868027 | 0.80 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr4_-_20092577 | 0.78 |
ENSDART00000164410
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr2_-_27996140 | 0.74 |
ENSDART00000097868
|
tgs1
|
trimethylguanosine synthase 1 |
| chr21_+_28710341 | 0.70 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr13_+_29794969 | 0.70 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr19_-_32178232 | 0.69 |
ENSDART00000103636
|
si:dkeyp-120h9.1
|
si:dkeyp-120h9.1 |
| chr1_-_53258987 | 0.68 |
ENSDART00000032552
|
taf5l
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr21_+_21242470 | 0.67 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr16_+_41004372 | 0.67 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr25_+_26458700 | 0.67 |
|
|
|
| chr13_+_28487504 | 0.65 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr12_+_5013049 | 0.63 |
ENSDART00000161548
|
kif22
|
kinesin family member 22 |
| chr25_-_10694465 | 0.61 |
ENSDART00000127054
|
BX572619.1
|
ENSDARG00000089303 |
| chr16_-_17678748 | 0.61 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr10_+_21487218 | 0.59 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr24_+_28304276 | 0.58 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr13_+_29794944 | 0.57 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr2_+_15379961 | 0.57 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr10_+_21487458 | 0.57 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr10_+_16914003 | 0.55 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr7_+_28341426 | 0.53 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr25_-_21666120 | 0.52 |
ENSDART00000089596
|
tmem168b
|
transmembrane protein 168b |
| chr11_+_5929498 | 0.51 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr19_-_32313943 | 0.51 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr19_+_1298795 | 0.51 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr16_+_41004516 | 0.50 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr22_-_52486 | 0.49 |
|
|
|
| chr21_+_18913636 | 0.49 |
ENSDART00000036172
|
crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr20_+_35156812 | 0.47 |
|
|
|
| chr17_+_6635602 | 0.46 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr7_-_48123296 | 0.46 |
ENSDART00000174381
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr2_+_15379717 | 0.46 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr12_-_34657387 | 0.45 |
ENSDART00000153418
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr22_-_52569 | 0.44 |
|
|
|
| chr21_-_18913374 | 0.44 |
|
|
|
| chr12_-_93105 | 0.44 |
ENSDART00000105694
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr20_-_44020387 | 0.43 |
ENSDART00000026213
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr3_-_21054281 | 0.43 |
ENSDART00000155550
|
taok2a
|
TAO kinase 2a |
| chr17_-_50140768 | 0.43 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr7_+_23224800 | 0.43 |
ENSDART00000115299
ENSDART00000101423 |
zgc:109889
|
zgc:109889 |
| chr21_+_28710424 | 0.43 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr9_+_8429767 | 0.43 |
ENSDART00000142796
|
zgc:153499
|
zgc:153499 |
| chr21_-_45589751 | 0.42 |
ENSDART00000164315
ENSDART00000164052 |
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr5_-_62371419 | 0.40 |
ENSDART00000161149
|
ENSDARG00000098860
|
ENSDARG00000098860 |
| chr20_-_44020837 | 0.40 |
ENSDART00000149897
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr7_+_23224924 | 0.39 |
ENSDART00000142401
|
zgc:109889
|
zgc:109889 |
| chr21_+_28710269 | 0.39 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr20_+_42350025 | 0.39 |
ENSDART00000084642
|
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr6_-_1315162 | 0.39 |
|
|
|
| chr19_-_32314264 | 0.39 |
|
|
|
| chr24_-_1014256 | 0.38 |
ENSDART00000114544
|
cdk13
|
cyclin-dependent kinase 13 |
| chr20_-_2708593 | 0.38 |
ENSDART00000145730
|
akirin2
|
akirin 2 |
| chr1_+_53409683 | 0.38 |
ENSDART00000108601
|
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr17_+_6635649 | 0.38 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr1_+_56632773 | 0.36 |
ENSDART00000104222
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr6_-_35326673 | 0.36 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr21_-_29995378 | 0.36 |
ENSDART00000178201
|
pwwp2a
|
PWWP domain containing 2A |
| chr11_-_25615683 | 0.36 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr5_-_47323844 | 0.35 |
ENSDART00000145665
ENSDART00000007057 |
ccnh
|
cyclin H |
| chr2_+_15380054 | 0.34 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr20_-_33584753 | 0.34 |
ENSDART00000061843
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr20_-_44020601 | 0.33 |
ENSDART00000149897
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr17_-_24848180 | 0.33 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr10_-_35377548 | 0.33 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr20_-_2708686 | 0.33 |
ENSDART00000145730
|
akirin2
|
akirin 2 |
| chr6_-_1315096 | 0.33 |
|
|
|
| chr16_+_23580507 | 0.33 |
ENSDART00000021092
|
snx27b
|
sorting nexin family member 27b |
| chr14_+_31149207 | 0.33 |
|
|
|
| chr5_-_37281096 | 0.32 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr22_-_52103 | 0.32 |
|
|
|
| chr11_-_21203166 | 0.32 |
ENSDART00000080051
|
RASSF5
|
Ras association domain family member 5 |
| chr11_+_269859 | 0.32 |
ENSDART00000109091
|
mettl1
|
methyltransferase like 1 |
| chr3_+_25719002 | 0.31 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr4_-_20092548 | 0.31 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr16_+_27629587 | 0.31 |
ENSDART00000111245
|
tmem67
|
transmembrane protein 67 |
| chr9_+_8429612 | 0.30 |
ENSDART00000178144
|
zgc:153499
|
zgc:153499 |
| chr11_-_21203114 | 0.30 |
ENSDART00000080051
|
RASSF5
|
Ras association domain family member 5 |
| chr20_+_42349759 | 0.29 |
ENSDART00000160519
ENSDART00000165631 |
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr9_+_1225877 | 0.27 |
ENSDART00000177730
|
CABZ01114359.1
|
ENSDARG00000107255 |
| chr24_-_1026977 | 0.25 |
ENSDART00000147508
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr3_+_25036636 | 0.25 |
ENSDART00000077493
|
TST
|
thiosulfate sulfurtransferase |
| chr19_+_5218562 | 0.24 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr8_+_49947734 | 0.24 |
ENSDART00000159816
ENSDART00000098707 |
naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr10_-_35377482 | 0.23 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr14_+_51724366 | 0.23 |
ENSDART00000168437
|
b4galt1l
|
DP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1, like |
| chr22_+_17278690 | 0.23 |
ENSDART00000088460
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr3_+_25718973 | 0.23 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr2_+_27996280 | 0.22 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr19_-_32314180 | 0.21 |
|
|
|
| chr9_+_24891584 | 0.21 |
|
|
|
| chr19_-_32313554 | 0.21 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr21_+_18913329 | 0.20 |
ENSDART00000036172
|
crkl
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr5_-_18457822 | 0.20 |
ENSDART00000142531
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr11_+_2641147 | 0.18 |
ENSDART00000132768
|
mapk14b
|
mitogen-activated protein kinase 14b |
| chr17_+_43633099 | 0.18 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr22_+_31110716 | 0.17 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr2_+_16505452 | 0.15 |
ENSDART00000045933
|
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr18_-_46371631 | 0.15 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| KN150266v1_-_69477 | 0.15 |
|
|
|
| chr8_+_21192955 | 0.13 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr24_+_17201072 | 0.12 |
ENSDART00000024722
|
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr5_+_47211006 | 0.12 |
ENSDART00000139824
|
rasa1a
|
RAS p21 protein activator (GTPase activating protein) 1a |
| chr23_+_2193828 | 0.11 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr22_-_37861903 | 0.11 |
ENSDART00000085931
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr12_+_18914438 | 0.10 |
ENSDART00000153086
|
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr12_-_92728 | 0.10 |
ENSDART00000152496
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr7_-_68961464 | 0.10 |
ENSDART00000168311
|
usp10
|
ubiquitin specific peptidase 10 |
| chr25_-_31496617 | 0.10 |
ENSDART00000155106
|
ENSDARG00000097988
|
ENSDARG00000097988 |
| chr20_+_28367799 | 0.10 |
ENSDART00000153319
ENSDART00000103330 |
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr5_+_30567648 | 0.08 |
ENSDART00000086443
|
atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr3_+_25036265 | 0.07 |
ENSDART00000077493
|
TST
|
thiosulfate sulfurtransferase |
| chr16_+_25891403 | 0.06 |
ENSDART00000154652
|
irgq1
|
immunity-related GTPase family, q1 |
| chr23_+_30781267 | 0.06 |
ENSDART00000016096
|
dnajc11a
|
DnaJ (Hsp40) homolog, subfamily C, member 11a |
| chr1_-_39414732 | 0.06 |
ENSDART00000135647
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr19_-_1376498 | 0.05 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr4_-_70726438 | 0.05 |
ENSDART00000124184
|
CABZ01054405.1
|
ENSDARG00000089604 |
| chr7_-_8715924 | 0.04 |
ENSDART00000111002
|
si:ch211-74f19.2
|
si:ch211-74f19.2 |
| chr19_+_8693914 | 0.04 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr7_-_64889963 | 0.04 |
ENSDART00000110614
ENSDART00000098277 ENSDART00000171493 |
tmem41b
|
transmembrane protein 41B |
| chr15_+_509915 | 0.03 |
|
|
|
| chr1_-_22905093 | 0.02 |
ENSDART00000138852
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr21_-_3619164 | 0.02 |
ENSDART00000133980
|
scamp1
|
secretory carrier membrane protein 1 |
| chr4_-_16324786 | 0.01 |
|
|
|
| chr22_-_20317131 | 0.01 |
ENSDART00000161610
|
tcf3b
|
transcription factor 3b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.2 | 0.8 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 3.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.7 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 1.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 3.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.4 | GO:0045639 | positive regulation of myeloid cell differentiation(GO:0045639) |
| 0.1 | 0.3 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 2.4 | GO:0045089 | positive regulation of defense response(GO:0031349) positive regulation of innate immune response(GO:0045089) |
| 0.1 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 1.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.8 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.4 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.3 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 3.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.0 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 3.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0030882 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 3.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |