Project
DANIO-CODE
Navigation
Downloads
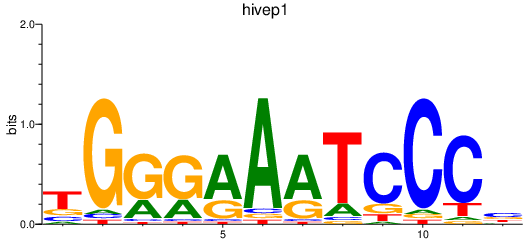
Results for hivep1
Z-value: 0.28
Transcription factors associated with hivep1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hivep1
|
ENSDARG00000103658 | HIVEP zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hivep1 | dr10_dc_chr19_-_3547521_3547630 | -0.33 | 2.1e-01 | Click! |
Activity profile of hivep1 motif
Sorted Z-values of hivep1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hivep1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_24971633 | 0.68 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr12_+_17632953 | 0.36 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr2_+_25106923 | 0.29 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr20_+_109649 | 0.28 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr16_+_1338291 | 0.24 |
ENSDART00000149299
|
cers2b
|
ceramide synthase 2b |
| chr16_+_46327528 | 0.22 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr7_-_60525897 | 0.20 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr13_+_4536343 | 0.20 |
|
|
|
| chr18_-_16934961 | 0.20 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr4_+_8007802 | 0.19 |
ENSDART00000014036
|
optn
|
optineurin |
| chr4_-_3184182 | 0.19 |
|
|
|
| chr20_-_42095643 | 0.19 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr10_+_37229202 | 0.19 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_+_19262722 | 0.18 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr6_-_6239268 | 0.18 |
|
|
|
| chr9_+_38778234 | 0.18 |
ENSDART00000131784
|
snx4
|
sorting nexin 4 |
| chr20_-_21775455 | 0.18 |
|
|
|
| chr17_-_20182801 | 0.17 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr25_+_16549708 | 0.17 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr6_+_54568388 | 0.16 |
ENSDART00000093199
|
tead3b
|
TEA domain family member 3 b |
| chr13_-_25689369 | 0.15 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr14_-_30747259 | 0.15 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr16_+_35582277 | 0.15 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr6_-_49863198 | 0.15 |
ENSDART00000121809
|
gnas
|
GNAS complex locus |
| chr13_+_22173410 | 0.15 |
ENSDART00000173405
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr7_+_31750808 | 0.15 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr22_+_3167234 | 0.14 |
ENSDART00000176409
|
ftsj3
|
FtsJ homolog 3 (E. coli) |
| chr2_-_51231808 | 0.14 |
ENSDART00000170470
|
ghrhra
|
growth hormone releasing hormone receptor a |
| chr13_-_2812369 | 0.13 |
|
|
|
| chr6_-_31380261 | 0.13 |
|
|
|
| chr11_+_269859 | 0.13 |
ENSDART00000109091
|
mettl1
|
methyltransferase like 1 |
| chr5_-_11074111 | 0.13 |
|
|
|
| chr25_+_3221696 | 0.13 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr9_-_28444933 | 0.13 |
|
|
|
| chr19_-_24971920 | 0.13 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr14_-_30747448 | 0.12 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr5_+_23582805 | 0.12 |
ENSDART00000137298
|
tp53
|
tumor protein p53 |
| chr22_-_26231606 | 0.12 |
ENSDART00000060888
|
ccdc130
|
coiled-coil domain containing 130 |
| chr11_-_19414624 | 0.12 |
ENSDART00000157847
|
atxn7
|
ataxin 7 |
| chr6_+_34491645 | 0.12 |
ENSDART00000178602
|
CR848703.1
|
ENSDARG00000106925 |
| chr11_-_15189966 | 0.12 |
|
|
|
| chr17_-_53241446 | 0.11 |
|
|
|
| chr11_+_26138359 | 0.11 |
ENSDART00000087652
|
cpne1
|
copine I |
| chr3_-_39316317 | 0.11 |
|
|
|
| chr1_-_674449 | 0.11 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr9_+_35205519 | 0.11 |
ENSDART00000100700
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr16_+_39321170 | 0.11 |
ENSDART00000141801
|
osbpl10b
|
oxysterol binding protein-like 10b |
| chr6_+_12810986 | 0.11 |
ENSDART00000104757
ENSDART00000165896 |
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr14_-_36004835 | 0.11 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr20_+_43794079 | 0.11 |
ENSDART00000045185
|
lin9
|
lin-9 DREAM MuvB core complex component |
| chr24_+_9316158 | 0.11 |
|
|
|
| chr9_+_33049445 | 0.10 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr19_+_43908885 | 0.10 |
ENSDART00000159421
|
BX649384.3
|
ENSDARG00000100839 |
| chr8_-_26917960 | 0.10 |
|
|
|
| chr19_+_22763741 | 0.10 |
ENSDART00000109157
|
fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr19_-_6856033 | 0.10 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr14_-_30747372 | 0.10 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr9_+_35205252 | 0.10 |
ENSDART00000100701
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr19_+_7630609 | 0.10 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr11_+_12662507 | 0.09 |
ENSDART00000135761
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr17_+_11519212 | 0.09 |
|
|
|
| chr25_+_24520476 | 0.09 |
|
|
|
| chr9_+_492585 | 0.09 |
ENSDART00000112635
|
ENSDARG00000078172
|
ENSDARG00000078172 |
| chr8_-_12394879 | 0.09 |
ENSDART00000101174
|
traf1
|
TNF receptor-associated factor 1 |
| chr4_-_16324786 | 0.09 |
|
|
|
| chr13_+_36832090 | 0.09 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr13_-_5953330 | 0.09 |
ENSDART00000099224
|
dld
|
deltaD |
| chr21_-_28629404 | 0.09 |
ENSDART00000125652
|
nrg2a
|
neuregulin 2a |
| chr3_+_35276575 | 0.09 |
ENSDART00000102994
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr15_-_16140187 | 0.09 |
ENSDART00000080413
|
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr12_-_25126122 | 0.09 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr20_+_23602537 | 0.08 |
|
|
|
| chr18_-_11707 | 0.08 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr13_+_36831946 | 0.08 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr19_+_25307436 | 0.08 |
ENSDART00000132209
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr7_-_667670 | 0.08 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr21_+_599565 | 0.08 |
ENSDART00000177311
|
CABZ01072933.1
|
ENSDARG00000106379 |
| chr10_-_7513764 | 0.08 |
ENSDART00000167054
ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr16_+_35582411 | 0.08 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr19_-_1921261 | 0.08 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr11_-_35757903 | 0.08 |
ENSDART00000167472
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr22_-_38964356 | 0.08 |
ENSDART00000131462
|
rbbp9
|
retinoblastoma binding protein 9 |
| chr22_-_20316792 | 0.08 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr8_-_18637019 | 0.07 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr2_+_50126491 | 0.07 |
ENSDART00000144060
|
rpl37
|
ribosomal protein L37 |
| chr23_-_33811895 | 0.07 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr2_-_9956527 | 0.07 |
|
|
|
| chr8_+_17493410 | 0.07 |
ENSDART00000147760
|
si:ch73-70k4.1
|
si:ch73-70k4.1 |
| chr3_+_32549515 | 0.07 |
ENSDART00000164428
|
kat8
|
K(lysine) acetyltransferase 8 |
| chr2_+_25107108 | 0.07 |
ENSDART00000136072
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr16_+_10666588 | 0.07 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr3_+_60988547 | 0.07 |
ENSDART00000161534
|
lmtk2
|
lemur tyrosine kinase 2 |
| chr14_+_50164568 | 0.07 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr10_-_42931440 | 0.06 |
ENSDART00000123608
|
zgc:100918
|
zgc:100918 |
| chr5_-_31257031 | 0.06 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr17_-_53241366 | 0.06 |
ENSDART00000171082
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr7_-_18751152 | 0.06 |
ENSDART00000157618
|
HACD4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr20_+_16982314 | 0.06 |
ENSDART00000146625
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr14_-_36004908 | 0.06 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr2_-_58788923 | 0.06 |
|
|
|
| KN150030v1_-_22613 | 0.06 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr22_+_3167367 | 0.06 |
ENSDART00000160604
|
ftsj3
|
FtsJ homolog 3 (E. coli) |
| chr7_-_60525749 | 0.06 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr19_-_42933592 | 0.06 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr19_-_3947422 | 0.06 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr20_+_13987091 | 0.06 |
ENSDART00000152611
|
nek2
|
NIMA-related kinase 2 |
| chr14_+_50164888 | 0.06 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr7_-_18751216 | 0.06 |
ENSDART00000157618
|
HACD4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr25_-_3221593 | 0.05 |
ENSDART00000082385
|
golt1bb
|
golgi transport 1Bb |
| chr3_+_35277256 | 0.05 |
ENSDART00000150918
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr13_-_24181106 | 0.05 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr8_-_18637047 | 0.05 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr13_-_5952897 | 0.05 |
ENSDART00000099224
|
dld
|
deltaD |
| chr11_+_12662882 | 0.05 |
ENSDART00000122812
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| KN150030v1_-_22642 | 0.05 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr7_+_37105282 | 0.05 |
|
|
|
| chr7_+_16583304 | 0.05 |
ENSDART00000113332
|
nav2a
|
neuron navigator 2a |
| chr11_-_36188531 | 0.05 |
ENSDART00000128889
|
zbtb40
|
zinc finger and BTB domain containing 40 |
| chr11_+_12662209 | 0.05 |
ENSDART00000054837
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_-_37419017 | 0.05 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr9_-_28444798 | 0.05 |
|
|
|
| chr7_+_57423271 | 0.05 |
ENSDART00000056466
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr3_+_56507117 | 0.05 |
ENSDART00000154405
|
rac1b
|
ras-related C3 botulinum toxin substrate 1b (rho family, small GTP binding protein Rac1) |
| chr22_+_30234718 | 0.04 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr12_+_28774063 | 0.04 |
ENSDART00000166229
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr7_-_667841 | 0.04 |
ENSDART00000160644
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr12_-_43310712 | 0.04 |
ENSDART00000178532
ENSDART00000179029 |
BX321882.2
|
ENSDARG00000109101 |
| chr3_+_34010958 | 0.04 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr14_+_50164534 | 0.04 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr2_-_37702092 | 0.04 |
ENSDART00000012191
|
hiat1a
|
hippocampus abundant transcript 1a |
| chr19_+_7798512 | 0.04 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr23_-_3815492 | 0.04 |
ENSDART00000131536
|
hmga1a
|
high mobility group AT-hook 1a |
| chr3_+_25036636 | 0.04 |
ENSDART00000077493
|
TST
|
thiosulfate sulfurtransferase |
| chr5_-_28594817 | 0.04 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr3_+_60988788 | 0.04 |
ENSDART00000110030
|
lmtk2
|
lemur tyrosine kinase 2 |
| chr25_-_8548210 | 0.04 |
ENSDART00000155280
|
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr5_+_36431907 | 0.04 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr10_+_10393377 | 0.04 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr16_+_6694341 | 0.04 |
|
|
|
| chr19_+_7630777 | 0.04 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr7_-_73625494 | 0.04 |
ENSDART00000041385
|
zgc:163061
|
zgc:163061 |
| chr7_+_16583234 | 0.03 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr19_-_7151801 | 0.03 |
ENSDART00000176808
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr14_+_8641470 | 0.03 |
ENSDART00000158025
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr25_-_8548290 | 0.03 |
ENSDART00000155280
|
GDPGP1
|
GDP-D-glucose phosphorylase 1 |
| chr16_+_39321120 | 0.03 |
ENSDART00000043823
|
osbpl10b
|
oxysterol binding protein-like 10b |
| chr3_-_39316355 | 0.03 |
|
|
|
| chr1_-_58606982 | 0.03 |
ENSDART00000158067
|
txndc11
|
thioredoxin domain containing 11 |
| chr20_-_42095478 | 0.03 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr21_+_599634 | 0.03 |
ENSDART00000177311
|
CABZ01072933.1
|
ENSDARG00000106379 |
| chr19_-_24971335 | 0.03 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr11_-_23460219 | 0.02 |
|
|
|
| chr11_-_38278666 | 0.02 |
|
|
|
| chr13_-_48465147 | 0.02 |
ENSDART00000132895
|
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| KN150108v1_-_8893 | 0.02 |
ENSDART00000167033
|
ENSDARG00000100513
|
ENSDARG00000100513 |
| chr18_-_40462054 | 0.02 |
|
|
|
| chr16_-_34330688 | 0.02 |
ENSDART00000044235
ENSDART00000159744 |
phactr4b
|
phosphatase and actin regulator 4b |
| chr16_+_10666906 | 0.02 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr25_+_3221887 | 0.02 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr14_-_36004765 | 0.02 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr21_+_3951807 | 0.02 |
ENSDART00000123759
|
rapgef1b
|
Rap guanine nucleotide exchange factor (GEF) 1b |
| chr7_-_6544898 | 0.02 |
ENSDART00000173270
ENSDART00000066450 |
dusp19b
|
dual specificity phosphatase 19b |
| chr5_-_28522443 | 0.02 |
ENSDART00000144802
|
dfnb31b
|
deafness, autosomal recessive 31b |
| chr6_-_51541332 | 0.02 |
ENSDART00000156336
ENSDART00000154512 |
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr9_-_52033978 | 0.02 |
ENSDART00000148918
|
tank
|
TRAF family member-associated NFKB activator |
| chr6_+_54568649 | 0.02 |
ENSDART00000157142
|
tead3b
|
TEA domain family member 3 b |
| chr12_-_4800362 | 0.01 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr22_+_30060679 | 0.01 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr13_+_36832145 | 0.01 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr10_-_42152265 | 0.01 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr4_+_73504354 | 0.01 |
ENSDART00000148575
|
nup50
|
nucleoporin 50 |
| chr19_-_12485032 | 0.01 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr7_-_667744 | 0.01 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr11_+_24826881 | 0.01 |
ENSDART00000157435
|
ndrg3a
|
ndrg family member 3a |
| chr6_-_113401 | 0.01 |
ENSDART00000151251
|
LRRC8E
|
leucine rich repeat containing 8 VRAC subunit E |
| chr15_+_17164373 | 0.01 |
ENSDART00000123197
ENSDART00000154418 |
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| KN150670v1_-_72156 | 0.01 |
|
|
|
| chr18_+_5637539 | 0.01 |
|
|
|
| chr22_-_20316908 | 0.01 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr24_-_20987184 | 0.01 |
ENSDART00000010126
|
zdhhc23b
|
zinc finger, DHHC-type containing 23b |
| chr5_+_29988157 | 0.01 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr13_+_40566900 | 0.01 |
ENSDART00000057047
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr14_+_22170431 | 0.01 |
ENSDART00000079409
|
nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr5_+_22076136 | 0.01 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr14_-_2114123 | 0.01 |
ENSDART00000111335
|
pcdh2g13
|
protocadherin 2 gamma 13 |
| chr19_+_7630730 | 0.01 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr11_+_12662284 | 0.01 |
ENSDART00000054837
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr6_-_6818925 | 0.01 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr6_-_6818607 | 0.01 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr5_-_60747054 | 0.01 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr6_-_39634534 | 0.01 |
ENSDART00000077839
|
atf7b
|
activating transcription factor 7b |
| chr22_-_26231695 | 0.01 |
ENSDART00000142821
|
ccdc130
|
coiled-coil domain containing 130 |
| chr5_-_31089598 | 0.00 |
ENSDART00000098172
|
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
| chr16_+_23172295 | 0.00 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr17_+_12788580 | 0.00 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr2_-_50126390 | 0.00 |
ENSDART00000083690
|
blvra
|
biliverdin reductase A |
| chr2_-_36691234 | 0.00 |
ENSDART00000140415
|
ccnl1b
|
cyclin L1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.1 | GO:0051701 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) interaction with host(GO:0051701) |
| 0.0 | 0.1 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.2 | GO:0035255 | beta-2 adrenergic receptor binding(GO:0031698) D1 dopamine receptor binding(GO:0031748) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |