Project
DANIO-CODE
Navigation
Downloads
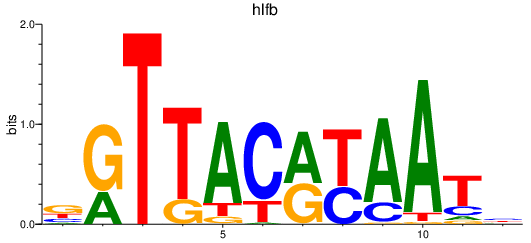
Results for hlfb
Z-value: 0.40
Transcription factors associated with hlfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hlfb
|
ENSDARG00000061011 | HLF transcription factor, PAR bZIP family member b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hlfb | dr10_dc_chr12_+_32044331_32044374 | -0.47 | 6.6e-02 | Click! |
Activity profile of hlfb motif
Sorted Z-values of hlfb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hlfb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_24617667 | 0.56 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr18_-_16933517 | 0.49 |
|
|
|
| chr6_+_50452131 | 0.42 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr6_+_16341787 | 0.40 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr16_+_53391438 | 0.38 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr1_-_35196892 | 0.35 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr22_+_15946773 | 0.34 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr13_-_10488076 | 0.33 |
ENSDART00000160561
|
si:ch73-54n14.2
|
si:ch73-54n14.2 |
| chr15_-_38027363 | 0.33 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr23_-_18972097 | 0.31 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr17_+_26946957 | 0.31 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr10_+_39140943 | 0.31 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr25_-_18234069 | 0.30 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr15_-_5912236 | 0.30 |
|
|
|
| chr21_-_34809442 | 0.29 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr15_-_29415309 | 0.29 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr6_-_26905146 | 0.28 |
ENSDART00000011863
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr5_+_6393799 | 0.26 |
ENSDART00000099417
|
CABZ01041962.1
|
ENSDARG00000076082 |
| chr13_-_35908743 | 0.26 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr24_+_38783264 | 0.26 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr6_+_16342020 | 0.26 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr17_-_20216303 | 0.26 |
ENSDART00000029380
|
bnip4
|
BCL2/adenovirus E1B interacting protein 4 |
| KN150442v1_-_37820 | 0.25 |
ENSDART00000158371
|
ptmab
|
prothymosin, alpha b |
| chr5_-_68439806 | 0.24 |
ENSDART00000168213
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr6_+_12091402 | 0.24 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr4_-_22617898 | 0.21 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr8_-_15071283 | 0.21 |
|
|
|
| chr10_+_32949485 | 0.21 |
|
|
|
| chr15_-_5911817 | 0.21 |
|
|
|
| chr15_-_5912105 | 0.21 |
|
|
|
| chr10_+_18919823 | 0.21 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr22_+_11745592 | 0.20 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr19_-_3115104 | 0.19 |
ENSDART00000093146
|
bop1
|
block of proliferation 1 |
| chr18_+_26916897 | 0.19 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr10_+_36718872 | 0.19 |
ENSDART00000063359
|
ucp3
|
uncoupling protein 3 |
| chr13_-_50940551 | 0.19 |
|
|
|
| chr8_+_7937128 | 0.19 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr21_+_26486084 | 0.19 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr4_-_16555765 | 0.18 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr6_+_48349630 | 0.18 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr7_+_30222432 | 0.18 |
ENSDART00000027466
|
fam63b
|
family with sequence similarity 63, member B |
| chr20_-_35343242 | 0.18 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr23_+_36608246 | 0.18 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| KN150702v1_+_177145 | 0.18 |
ENSDART00000166225
|
CABZ01076674.1
|
ENSDARG00000102620 |
| chr23_-_32239909 | 0.18 |
|
|
|
| chr24_-_9153377 | 0.17 |
ENSDART00000082434
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr8_-_23172938 | 0.17 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr8_+_7937320 | 0.17 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr22_-_35967525 | 0.17 |
ENSDART00000176971
|
CABZ01037120.1
|
ENSDARG00000106725 |
| chr6_+_42478185 | 0.17 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr20_-_36714338 | 0.17 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr14_-_30564987 | 0.17 |
ENSDART00000173451
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr1_-_11707244 | 0.16 |
ENSDART00000146067
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr24_-_16878970 | 0.16 |
ENSDART00000106058
ENSDART00000106057 |
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr10_+_16914003 | 0.15 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr16_+_53391776 | 0.15 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr1_-_39177682 | 0.15 |
ENSDART00000035739
|
tmem134
|
transmembrane protein 134 |
| chr21_+_18870471 | 0.15 |
ENSDART00000160185
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr21_+_17014240 | 0.15 |
ENSDART00000047201
|
atp2a2b
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2b |
| chr21_-_32747483 | 0.15 |
|
|
|
| chr19_-_22914516 | 0.15 |
|
|
|
| chr6_+_23709605 | 0.15 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr16_-_45950530 | 0.14 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr7_+_45747622 | 0.14 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr21_-_32747592 | 0.14 |
|
|
|
| chr16_-_19762541 | 0.14 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr9_-_9754075 | 0.14 |
ENSDART00000092884
|
lrrc58b
|
leucine rich repeat containing 58b |
| chr12_-_26445560 | 0.14 |
ENSDART00000138437
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr21_+_25588388 | 0.14 |
ENSDART00000134678
|
ENSDARG00000078256
|
ENSDARG00000078256 |
| chr24_+_28481885 | 0.14 |
ENSDART00000079770
ENSDART00000147063 |
abca4a
|
ATP-binding cassette, sub-family A (ABC1), member 4a |
| chr6_-_32425011 | 0.13 |
ENSDART00000078908
|
usp1
|
ubiquitin specific peptidase 1 |
| chr3_+_26092485 | 0.13 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr5_+_22470117 | 0.13 |
ENSDART00000020434
|
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr19_+_4939815 | 0.13 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr7_+_45748086 | 0.13 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr20_-_36714401 | 0.13 |
ENSDART00000165176
|
enah
|
enabled homolog (Drosophila) |
| chr19_+_12066023 | 0.13 |
ENSDART00000130537
|
spag1a
|
sperm associated antigen 1a |
| chr13_-_18064929 | 0.13 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr4_+_22146112 | 0.13 |
ENSDART00000158316
|
acss3
|
acyl-CoA synthetase short-chain family member 3 |
| chr9_-_32489944 | 0.13 |
ENSDART00000078596
|
hspd1
|
heat shock 60 protein 1 |
| chr19_-_44459352 | 0.13 |
ENSDART00000151084
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr14_+_6656015 | 0.13 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr3_-_29760552 | 0.13 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| KN150708v1_-_31573 | 0.13 |
ENSDART00000160040
|
slc16a6a
|
solute carrier family 16, member 6a |
| chr11_-_44724371 | 0.13 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr5_+_18837448 | 0.13 |
|
|
|
| chr3_+_23963926 | 0.13 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr3_+_24564249 | 0.12 |
ENSDART00000111769
|
mkl1a
|
megakaryoblastic leukemia (translocation) 1a |
| chr24_-_38782898 | 0.12 |
ENSDART00000155542
|
FP102170.1
|
ENSDARG00000097123 |
| chr5_-_11309164 | 0.12 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr22_+_11726312 | 0.12 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr19_+_16111610 | 0.12 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr7_+_22582619 | 0.12 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
| chr13_-_50940520 | 0.12 |
|
|
|
| chr3_+_7877162 | 0.12 |
ENSDART00000057434
ENSDART00000170291 |
hook2
|
hook microtubule-tethering protein 2 |
| chr13_-_35908796 | 0.12 |
ENSDART00000084929
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr6_+_49552893 | 0.12 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr4_-_16556116 | 0.12 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr2_+_24152953 | 0.12 |
ENSDART00000131030
|
gorasp1a
|
golgi reassembly stacking protein 1a |
| chr15_-_47277391 | 0.12 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr23_-_14460361 | 0.12 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr18_-_14910429 | 0.11 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr16_-_6909034 | 0.11 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr19_-_22914419 | 0.11 |
|
|
|
| chr21_-_32748287 | 0.11 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr24_+_37911828 | 0.11 |
ENSDART00000131975
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr10_-_5016621 | 0.11 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr25_+_34340569 | 0.11 |
ENSDART00000157638
|
tmem231
|
transmembrane protein 231 |
| chr8_-_53165501 | 0.11 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr2_+_21698627 | 0.11 |
ENSDART00000133228
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr22_-_12305110 | 0.11 |
ENSDART00000123574
|
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr19_+_42658273 | 0.11 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr10_-_2848524 | 0.11 |
ENSDART00000034555
ENSDART00000135708 ENSDART00000137180 |
ddx56
|
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr7_+_45747395 | 0.11 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr25_-_6262259 | 0.11 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr7_+_45748011 | 0.10 |
ENSDART00000170294
|
ccne1
|
cyclin E1 |
| chr16_-_45943282 | 0.10 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr12_+_26971687 | 0.10 |
|
|
|
| chr24_-_9153339 | 0.10 |
ENSDART00000082434
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr5_-_13872772 | 0.10 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr20_-_19632944 | 0.10 |
ENSDART00000125830
|
eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr11_-_22211478 | 0.10 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr6_-_32424706 | 0.10 |
ENSDART00000078908
|
usp1
|
ubiquitin specific peptidase 1 |
| chr24_+_32610480 | 0.10 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr5_-_11309124 | 0.10 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr21_-_32747544 | 0.10 |
|
|
|
| chr14_+_37203442 | 0.10 |
|
|
|
| chr12_+_13167781 | 0.10 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr24_-_6304289 | 0.10 |
ENSDART00000140212
|
zgc:174877
|
zgc:174877 |
| chr22_-_15567180 | 0.10 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr21_+_4345107 | 0.09 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr9_-_9754243 | 0.09 |
ENSDART00000092884
|
lrrc58b
|
leucine rich repeat containing 58b |
| chr2_-_50126390 | 0.09 |
ENSDART00000083690
|
blvra
|
biliverdin reductase A |
| chr7_-_35136743 | 0.09 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr15_+_28370102 | 0.09 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr3_-_30754236 | 0.09 |
ENSDART00000109104
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr15_-_1800139 | 0.09 |
ENSDART00000093074
|
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr3_-_57459668 | 0.09 |
ENSDART00000003066
ENSDART00000125439 |
cyth1a
|
cytohesin 1a |
| chr7_-_37283707 | 0.09 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr1_-_49967146 | 0.09 |
ENSDART00000015591
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr9_+_27909662 | 0.09 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr2_+_16928379 | 0.09 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr20_-_7303735 | 0.09 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr2_+_23196172 | 0.09 |
ENSDART00000145944
|
amotl2b
|
angiomotin like 2b |
| chr23_-_29952020 | 0.09 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr3_+_16514376 | 0.09 |
ENSDART00000013816
|
ENSDARG00000013161
|
ENSDARG00000013161 |
| chr18_+_5986137 | 0.09 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr12_-_23008264 | 0.09 |
ENSDART00000152887
|
armc4
|
armadillo repeat containing 4 |
| chr5_-_19691538 | 0.09 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr8_-_8659980 | 0.09 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr21_-_4842528 | 0.09 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr18_-_14910358 | 0.09 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr25_+_13695348 | 0.09 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr12_+_19159234 | 0.09 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr22_+_10577272 | 0.09 |
ENSDART00000147975
|
rad54l2
|
RAD54-like 2 (S. cerevisiae) |
| chr3_-_40407003 | 0.09 |
|
|
|
| chr12_+_31307634 | 0.09 |
ENSDART00000153179
|
zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr23_+_4773894 | 0.09 |
ENSDART00000172739
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr5_+_2440622 | 0.09 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr13_+_12651778 | 0.09 |
ENSDART00000090000
|
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr16_+_17706003 | 0.09 |
|
|
|
| chr17_-_22552999 | 0.08 |
ENSDART00000141523
|
exo1
|
exonuclease 1 |
| chr5_-_13872815 | 0.08 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr9_-_306569 | 0.08 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr4_-_11978689 | 0.08 |
|
|
|
| chr4_-_25807510 | 0.08 |
ENSDART00000122881
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr13_-_14798134 | 0.08 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr11_+_24583090 | 0.08 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr19_+_12730277 | 0.08 |
ENSDART00000088917
|
rnmt
|
RNA (guanine-7-) methyltransferase |
| chr23_-_39879096 | 0.08 |
ENSDART00000159519
|
ENSDARG00000102199
|
ENSDARG00000102199 |
| chr25_+_34340352 | 0.08 |
ENSDART00000163190
|
tmem231
|
transmembrane protein 231 |
| chr5_+_35839198 | 0.08 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr22_+_15947127 | 0.08 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr13_-_36486255 | 0.08 |
ENSDART00000160250
|
BX927407.1
|
ENSDARG00000103360 |
| chr10_+_23130414 | 0.08 |
ENSDART00000022170
|
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr5_+_22470177 | 0.08 |
ENSDART00000020434
|
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr8_-_37217197 | 0.08 |
ENSDART00000178556
|
rbm39b
|
RNA binding motif protein 39b |
| chr7_+_22582561 | 0.08 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
| chr12_-_26445609 | 0.08 |
ENSDART00000132737
ENSDART00000163931 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr18_-_14910497 | 0.08 |
ENSDART00000099701
|
selo
|
selenoprotein O |
| chr1_+_15827083 | 0.08 |
|
|
|
| chr12_-_22279713 | 0.08 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr13_-_25066650 | 0.08 |
|
|
|
| chr13_+_29904913 | 0.08 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr17_-_10682357 | 0.08 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr8_-_22537415 | 0.08 |
ENSDART00000165640
|
porcnl
|
porcupine homolog like |
| chr11_-_7068532 | 0.08 |
ENSDART00000112156
|
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr7_-_21620707 | 0.07 |
ENSDART00000019699
|
mettl3
|
methyltransferase like 3 |
| chr24_+_26183653 | 0.07 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr22_+_12306260 | 0.07 |
|
|
|
| chr5_-_4507692 | 0.07 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr12_+_13167278 | 0.07 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr11_+_16017857 | 0.07 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr14_-_31222400 | 0.07 |
ENSDART00000034979
|
mospd1
|
motile sperm domain containing 1 |
| chr18_-_16934756 | 0.07 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr10_+_6925373 | 0.07 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr25_+_34340139 | 0.07 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr24_+_32610430 | 0.07 |
ENSDART00000132417
|
yme1l1a
|
YME1-like 1a |
| chr20_-_33584483 | 0.07 |
ENSDART00000177645
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr7_-_19390325 | 0.07 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.3 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.1 | 0.3 | GO:0003433 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0045041 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0090365 | forebrain radial glial cell differentiation(GO:0021861) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.0 | 0.1 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.4 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 0.1 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.2 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0035552 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic cell cycle, embryonic(GO:0045448) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.3 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.1 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.0 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.1 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.1 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.1 | GO:0097345 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.0 | GO:0034552 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016422 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |