Project
DANIO-CODE
Navigation
Downloads
Results for hlx1
Z-value: 0.40
Transcription factors associated with hlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hlx1
|
ENSDARG00000009134 | H2.0-like homeo box 1 (Drosophila) |
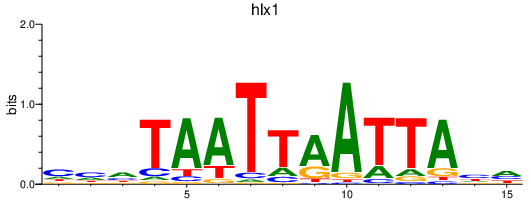
Activity profile of hlx1 motif
Sorted Z-values of hlx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hlx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_28441951 | 1.06 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr9_-_23307419 | 0.95 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr16_+_46435014 | 0.87 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr10_-_22948684 | 0.78 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr17_-_29102320 | 0.77 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr17_+_25500291 | 0.65 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr1_-_25600988 | 0.63 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr13_-_31491759 | 0.60 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr1_-_18809429 | 0.55 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr21_-_30508374 | 0.55 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr18_-_7184699 | 0.42 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr18_+_22367874 | 0.34 |
|
|
|
| chr7_+_21006803 | 0.25 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr2_+_29729161 | 0.22 |
ENSDART00000141666
|
AL845362.1
|
ENSDARG00000093205 |
| chr14_-_6901415 | 0.22 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr2_-_10020770 | 0.22 |
ENSDART00000046587
|
ap2m1a
|
adaptor-related protein complex 2, mu 1 subunit, a |
| chr16_+_37519702 | 0.21 |
|
|
|
| chr4_-_22617898 | 0.19 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr17_-_29102271 | 0.17 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr17_+_49198580 | 0.12 |
ENSDART00000059255
ENSDART00000155599 |
zgc:113176
|
zgc:113176 |
| chr2_+_1881022 | 0.11 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr21_-_27584637 | 0.03 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.9 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0007338 | single fertilization(GO:0007338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |