Project
DANIO-CODE
Navigation
Downloads
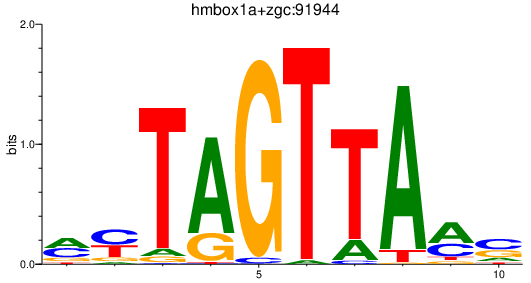
Results for hmbox1a+zgc:91944
Z-value: 0.78
Transcription factors associated with hmbox1a+zgc:91944
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmbox1a
|
ENSDARG00000027082 | homeobox containing 1a |
|
zgc
|
ENSDARG00000035887 | 91944 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zgc:91944 | dr10_dc_chr19_-_33013748_33013780 | -0.66 | 5.5e-03 | Click! |
| hmbox1a | dr10_dc_chr17_-_16316442_16316465 | 0.42 | 1.1e-01 | Click! |
Activity profile of hmbox1a+zgc:91944 motif
Sorted Z-values of hmbox1a+zgc:91944 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hmbox1a+zgc:91944
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22222388 | 1.55 |
ENSDART00000154376
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr2_-_21780380 | 1.50 |
ENSDART00000144587
|
plcd1b
|
phospholipase C, delta 1b |
| chr1_-_44893257 | 1.47 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_+_31997954 | 1.38 |
ENSDART00000164108
|
gmnn
|
geminin, DNA replication inhibitor |
| chr5_-_19619115 | 1.34 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr1_+_29919378 | 1.21 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr14_+_44703094 | 1.16 |
|
|
|
| chr6_-_53049946 | 1.13 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr23_-_45107021 | 1.07 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr5_-_34364552 | 1.03 |
ENSDART00000133170
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr17_+_1063519 | 1.01 |
|
|
|
| chr17_-_7661762 | 1.00 |
ENSDART00000135538
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr5_-_14783255 | 1.00 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr1_-_28257339 | 0.98 |
ENSDART00000148331
|
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr12_+_4650974 | 0.98 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr5_-_11309164 | 0.97 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr1_-_43347290 | 0.94 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr19_+_27056002 | 0.89 |
ENSDART00000175951
|
ints3
|
integrator complex subunit 3 |
| chr9_+_38832959 | 0.86 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr9_-_56692366 | 0.84 |
|
|
|
| chr6_+_44199738 | 0.83 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr9_+_22846286 | 0.83 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr7_+_36195907 | 0.82 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr13_-_21541531 | 0.80 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr2_+_223139 | 0.80 |
ENSDART00000113021
|
dhx30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr5_+_33949157 | 0.79 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr8_-_45932088 | 0.78 |
ENSDART00000040066
ENSDART00000132297 |
adam9
|
ADAM metallopeptidase domain 9 |
| chr3_+_18691103 | 0.77 |
ENSDART00000128626
|
tmem104
|
transmembrane protein 104 |
| chr2_+_24046083 | 0.77 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr1_-_43347143 | 0.76 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr2_+_24045890 | 0.76 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr17_+_28607470 | 0.76 |
ENSDART00000109693
|
heatr5a
|
HEAT repeat containing 5a |
| chr19_+_30210491 | 0.75 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr3_-_55252907 | 0.74 |
ENSDART00000108995
|
tex2
|
testis expressed 2 |
| chr14_-_23980060 | 0.73 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr5_-_11309124 | 0.72 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr3_-_23919508 | 0.71 |
ENSDART00000155896
|
BX936342.1
|
ENSDARG00000097621 |
| chr17_+_31722625 | 0.68 |
ENSDART00000156180
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr15_+_6462447 | 0.68 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr9_-_56423275 | 0.67 |
|
|
|
| chr13_+_35346924 | 0.67 |
ENSDART00000011583
|
mkks
|
McKusick-Kaufman syndrome |
| chr20_-_5992604 | 0.67 |
ENSDART00000170468
|
cep128
|
centrosomal protein 128 |
| chr16_-_43061368 | 0.66 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr1_+_43694707 | 0.65 |
ENSDART00000159686
|
unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr16_-_43107682 | 0.65 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr16_+_29756032 | 0.64 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr13_-_21529695 | 0.64 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr2_-_41888389 | 0.64 |
|
|
|
| chr7_+_36196295 | 0.64 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr19_-_22761674 | 0.64 |
|
|
|
| chr9_+_8919663 | 0.63 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr1_-_40536800 | 0.63 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr5_-_53777415 | 0.63 |
ENSDART00000169270
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr24_-_10935904 | 0.63 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr13_+_28655338 | 0.61 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr18_-_30043070 | 0.61 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr13_+_28655433 | 0.61 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr20_-_40589999 | 0.60 |
ENSDART00000075070
|
hsf2
|
heat shock transcription factor 2 |
| chr12_+_33257120 | 0.60 |
|
|
|
| chr3_+_48811632 | 0.59 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr6_-_30696540 | 0.58 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr19_+_12487051 | 0.58 |
ENSDART00000052238
ENSDART00000013865 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr11_+_24919694 | 0.58 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr11_-_35968053 | 0.58 |
ENSDART00000135888
|
rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr5_-_7418398 | 0.58 |
|
|
|
| chr7_-_24862634 | 0.57 |
ENSDART00000173781
|
badb
|
BCL2-associated agonist of cell death b |
| chr24_+_21531899 | 0.57 |
ENSDART00000158833
|
si:ch211-140b10.6
|
si:ch211-140b10.6 |
| chr17_+_28607337 | 0.56 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr3_+_58417512 | 0.55 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr14_+_8136545 | 0.55 |
ENSDART00000163059
|
si:dkey-160o24.3
|
si:dkey-160o24.3 |
| chr7_-_38363533 | 0.55 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr21_-_36768113 | 0.54 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr21_-_2170199 | 0.54 |
|
|
|
| chr8_-_22252628 | 0.53 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr7_+_34919736 | 0.53 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr9_-_25517542 | 0.53 |
ENSDART00000060840
|
med4
|
mediator complex subunit 4 |
| chr18_+_17552191 | 0.52 |
|
|
|
| chr2_-_52156537 | 0.52 |
ENSDART00000168875
|
si:ch211-9d9.1
|
si:ch211-9d9.1 |
| chr14_-_36096895 | 0.52 |
ENSDART00000173350
|
aga
|
aspartylglucosaminidase |
| chr13_-_35924447 | 0.51 |
ENSDART00000134955
|
lgmn
|
legumain |
| chr4_-_4498775 | 0.51 |
ENSDART00000130588
|
tbc1d30
|
TBC1 domain family, member 30 |
| chr16_+_54350976 | 0.51 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr3_-_34483494 | 0.50 |
ENSDART00000113036
|
zgc:163143
|
zgc:163143 |
| chr13_-_402878 | 0.50 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr6_-_40009230 | 0.50 |
ENSDART00000085666
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr24_-_24884729 | 0.50 |
ENSDART00000168254
|
nup58
|
nucleoporin 58 |
| chr7_+_8834138 | 0.50 |
ENSDART00000173250
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr19_+_44073792 | 0.50 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr19_+_27055869 | 0.49 |
ENSDART00000067793
ENSDART00000124755 |
ints3
|
integrator complex subunit 3 |
| chr15_+_23598869 | 0.48 |
ENSDART00000152320
|
si:dkey-182i3.10
|
si:dkey-182i3.10 |
| chr9_-_56692281 | 0.48 |
|
|
|
| chr6_+_59580554 | 0.48 |
|
|
|
| chr13_+_46429054 | 0.48 |
ENSDART00000160401
|
tmem63ba
|
transmembrane protein 63Ba |
| chr10_-_44173041 | 0.48 |
|
|
|
| chr10_+_22065599 | 0.48 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr16_-_32254735 | 0.48 |
ENSDART00000023995
|
fam26e.2
|
family with sequence similarity 26, member E, tandem duplicate 2 |
| chr7_+_33965084 | 0.47 |
ENSDART00000052474
|
tipin
|
timeless interacting protein |
| chr5_-_28959646 | 0.47 |
ENSDART00000133949
ENSDART00000167115 ENSDART00000051433 |
nelfb
|
negative elongation factor complex member B |
| chr23_+_25942056 | 0.47 |
ENSDART00000103934
|
ttpal
|
tocopherol (alpha) transfer protein-like |
| chr14_-_16118404 | 0.46 |
|
|
|
| chr12_+_10668405 | 0.46 |
ENSDART00000161986
ENSDART00000158227 |
top2a
|
topoisomerase (DNA) II alpha |
| chr11_+_23799984 | 0.45 |
|
|
|
| chr9_+_27518890 | 0.44 |
ENSDART00000111039
|
GTPBP8
|
GTP binding protein 8 (putative) |
| chr14_+_23419864 | 0.44 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr1_-_43347586 | 0.44 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr25_-_24440001 | 0.44 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr13_-_31807963 | 0.43 |
ENSDART00000026726
|
diexf
|
digestive organ expansion factor homolog |
| chr9_+_28778094 | 0.43 |
|
|
|
| chr18_-_18595722 | 0.43 |
ENSDART00000159274
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr18_+_22426900 | 0.43 |
ENSDART00000115388
|
cfdp1
|
craniofacial development protein 1 |
| chr1_+_40428827 | 0.43 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr9_-_19358693 | 0.43 |
ENSDART00000099396
|
zgc:152951
|
zgc:152951 |
| chr19_+_46371192 | 0.42 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr1_-_1780311 | 0.42 |
ENSDART00000135193
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr19_+_44073707 | 0.41 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr13_+_28655188 | 0.41 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr17_+_24090741 | 0.40 |
ENSDART00000064067
|
ehbp1
|
EH domain binding protein 1 |
| chr15_-_3921649 | 0.40 |
ENSDART00000157583
|
rnf168
|
ring finger protein 168 |
| chr2_+_24045941 | 0.40 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr8_+_31325461 | 0.39 |
|
|
|
| chr25_+_16259634 | 0.39 |
ENSDART00000136454
|
tead1a
|
TEA domain family member 1a |
| chr2_-_21967384 | 0.39 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr11_-_40239946 | 0.38 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr3_-_27159896 | 0.38 |
ENSDART00000151742
|
BX908803.1
|
ENSDARG00000096260 |
| chr20_-_31035087 | 0.37 |
ENSDART00000147359
|
wtap
|
WT1 associated protein |
| chr4_+_13453584 | 0.37 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr2_-_47827343 | 0.37 |
ENSDART00000056882
|
cul3a
|
cullin 3a |
| chr14_-_5509554 | 0.37 |
|
|
|
| chr7_-_5172270 | 0.37 |
ENSDART00000172963
|
FP236789.1
|
ENSDARG00000105406 |
| chr12_+_19159234 | 0.36 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr25_-_19388381 | 0.36 |
ENSDART00000154986
|
zgc:193812
|
zgc:193812 |
| chr17_-_14443647 | 0.36 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8-associated membrane protein |
| chr10_-_35377548 | 0.36 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr6_+_37463920 | 0.36 |
ENSDART00000086732
|
gtpbp6
|
GTP binding protein 6 (putative) |
| chr7_-_24778134 | 0.35 |
ENSDART00000135179
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr20_-_30132596 | 0.35 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr12_+_4651037 | 0.35 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr15_+_23588063 | 0.34 |
ENSDART00000152517
|
si:dkey-182i3.9
|
si:dkey-182i3.9 |
| chr23_+_44635770 | 0.34 |
|
|
|
| chr14_+_623802 | 0.34 |
ENSDART00000169624
|
zgc:158257
|
zgc:158257 |
| chr3_+_32520272 | 0.34 |
ENSDART00000164335
|
thoc6
|
THO complex 6 |
| chr11_+_12662882 | 0.34 |
ENSDART00000122812
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr8_+_19589684 | 0.34 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr3_+_24510974 | 0.34 |
ENSDART00000148414
ENSDART00000055590 |
zgc:113411
|
zgc:113411 |
| chr15_+_17138134 | 0.33 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr25_-_12838540 | 0.33 |
ENSDART00000167362
|
ccl39.7
|
chemokine (C-C motif) ligand 39, duplicate 7 |
| chr12_+_19159391 | 0.33 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr16_+_26864804 | 0.33 |
ENSDART00000163110
|
BX511010.2
|
ENSDARG00000101437 |
| chr19_-_7114660 | 0.33 |
ENSDART00000124094
|
daxx
|
death-domain associated protein |
| chr10_+_42111560 | 0.33 |
ENSDART00000126248
|
tmem120b
|
transmembrane protein 120B |
| chr3_+_58417635 | 0.32 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr2_-_21967353 | 0.32 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr19_+_31998135 | 0.32 |
ENSDART00000164108
|
gmnn
|
geminin, DNA replication inhibitor |
| chr14_+_23419894 | 0.31 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr14_-_38441710 | 0.31 |
|
|
|
| chr15_-_34799968 | 0.31 |
ENSDART00000110964
|
bag6
|
BCL2-associated athanogene 6 |
| chr14_+_38505984 | 0.30 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr2_+_10362661 | 0.30 |
ENSDART00000136018
|
nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr12_-_27496942 | 0.30 |
ENSDART00000066282
|
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr13_+_38688704 | 0.30 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr7_+_52477995 | 0.30 |
ENSDART00000053814
|
mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr12_-_31619559 | 0.30 |
|
|
|
| chr9_+_29832294 | 0.29 |
ENSDART00000023210
|
trim13
|
tripartite motif containing 13 |
| chr8_+_7815256 | 0.29 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr3_+_32519965 | 0.28 |
ENSDART00000131850
ENSDART00000055338 |
thoc6
|
THO complex 6 |
| chr20_+_31375056 | 0.28 |
|
|
|
| chr7_+_73583860 | 0.28 |
ENSDART00000166244
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr5_-_53776769 | 0.28 |
ENSDART00000164595
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr24_+_15510787 | 0.27 |
ENSDART00000042943
|
fbxo15
|
F-box protein 15 |
| chr18_-_31073514 | 0.27 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr7_-_61693560 | 0.27 |
ENSDART00000062704
|
plaa
|
phospholipase A2-activating protein |
| chr6_-_53049894 | 0.27 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr3_+_22204475 | 0.27 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr7_+_27531184 | 0.27 |
ENSDART00000079042
|
copb1
|
coatomer protein complex, subunit beta 1 |
| chr12_-_27497159 | 0.27 |
ENSDART00000066282
|
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr1_+_29919605 | 0.27 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr13_-_402908 | 0.26 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr9_-_25517664 | 0.26 |
ENSDART00000060840
|
med4
|
mediator complex subunit 4 |
| chr9_-_21572686 | 0.26 |
ENSDART00000038923
|
ift88
|
intraflagellar transport 88 homolog |
| chr3_+_22204569 | 0.26 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr14_+_15846060 | 0.26 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr25_-_31049064 | 0.25 |
ENSDART00000009126
|
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr7_+_5782298 | 0.25 |
ENSDART00000110632
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr14_+_38505790 | 0.25 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr14_-_28228482 | 0.25 |
ENSDART00000166611
|
zgc:113364
|
zgc:113364 |
| chr13_+_43739741 | 0.24 |
ENSDART00000174769
|
CU138544.1
|
ENSDARG00000108100 |
| chr3_+_54551887 | 0.24 |
ENSDART00000169663
|
CT573860.1
|
ENSDARG00000098327 |
| chr16_+_45964212 | 0.24 |
ENSDART00000128840
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr2_-_42279072 | 0.24 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr3_+_54507075 | 0.24 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr11_+_24919823 | 0.24 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr14_-_16118606 | 0.23 |
|
|
|
| chr20_-_30132167 | 0.23 |
ENSDART00000033588
|
BX248324.1
|
ENSDARG00000024870 |
| chr13_-_44715398 | 0.23 |
ENSDART00000136455
|
si:dkeyp-2e4.2
|
si:dkeyp-2e4.2 |
| chr13_-_35716712 | 0.23 |
ENSDART00000170106
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr12_-_13611954 | 0.23 |
ENSDART00000124638
ENSDART00000124364 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr2_+_10362753 | 0.23 |
ENSDART00000136018
|
nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr14_+_38506051 | 0.22 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr19_+_46371377 | 0.22 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr19_+_46371722 | 0.22 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.6 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.6 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 0.5 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.2 | 1.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 0.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.7 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 1.5 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 2.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.5 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.5 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 0.4 | GO:0010985 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.4 | GO:0045190 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.1 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.4 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 0.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.6 | GO:0034121 | regulation of toll-like receptor signaling pathway(GO:0034121) |
| 0.1 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.7 | GO:0097324 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 1.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.5 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.3 | GO:0034341 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) cellular response to interleukin-1(GO:0071347) |
| 0.1 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.2 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 1.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.9 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0006265 | resolution of meiotic recombination intermediates(GO:0000712) DNA topological change(GO:0006265) |
| 0.0 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 1.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.8 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.0 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 0.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 0.5 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 1.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.5 | GO:0003948 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity(GO:0003948) |
| 0.1 | 1.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.9 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.9 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.7 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |