Project
DANIO-CODE
Navigation
Downloads
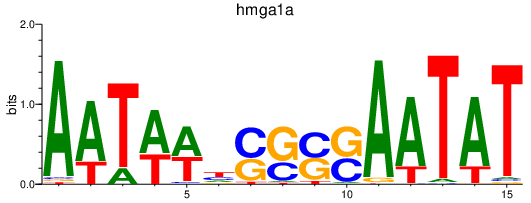
Results for hmga1a
Z-value: 1.06
Transcription factors associated with hmga1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmga1a
|
ENSDARG00000028335 | high mobility group AT-hook 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmga1a | dr10_dc_chr23_-_3815492_3815556 | 0.56 | 2.5e-02 | Click! |
Activity profile of hmga1a motif
Sorted Z-values of hmga1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hmga1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_25428372 | 2.88 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr23_-_28420470 | 2.27 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr7_-_22685672 | 2.22 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr3_+_61924544 | 2.22 |
ENSDART00000090370
|
noxo1a
|
NADPH oxidase organizer 1a |
| chr8_-_50992034 | 2.14 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr4_+_7406743 | 2.04 |
ENSDART00000168327
|
cald1a
|
caldesmon 1a |
| chr12_-_650506 | 2.04 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr5_-_55843553 | 1.95 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr25_+_33833810 | 1.92 |
ENSDART00000073440
|
ENSDARG00000051762
|
ENSDARG00000051762 |
| chr17_+_51430928 | 1.82 |
ENSDART00000170951
|
PXDN
|
peroxidasin |
| chr11_-_21422588 | 1.70 |
|
|
|
| chr8_+_53173227 | 1.50 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr4_-_978592 | 1.44 |
ENSDART00000160902
|
naga
|
N-acetylgalactosaminidase, alpha |
| chr20_+_34497817 | 1.44 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr21_-_30144979 | 1.39 |
ENSDART00000065447
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr3_+_39821041 | 1.38 |
|
|
|
| chr11_-_16840339 | 1.34 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr21_+_22808694 | 1.32 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr12_-_10372055 | 1.31 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr5_-_20310793 | 1.28 |
ENSDART00000147639
|
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr19_-_46058954 | 1.24 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr5_-_37850885 | 1.23 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr25_+_558191 | 1.18 |
ENSDART00000126863
ENSDART00000166012 |
nell2a
|
neural EGFL like 2a |
| chr5_-_65103730 | 1.12 |
ENSDART00000169616
|
notch1b
|
notch 1b |
| chr10_+_42905246 | 1.11 |
|
|
|
| chr6_-_48095609 | 1.07 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr23_-_45646407 | 1.03 |
ENSDART00000149291
|
ENSDARG00000036894
|
ENSDARG00000036894 |
| chr2_+_1055716 | 1.00 |
ENSDART00000165477
|
AL935300.3
|
ENSDARG00000098661 |
| chr7_-_22685631 | 0.96 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr16_-_12345194 | 0.95 |
ENSDART00000114759
|
lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr8_-_45043860 | 0.93 |
ENSDART00000138269
|
BX005065.1
|
ENSDARG00000092243 |
| chr18_+_14627030 | 0.93 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr17_-_50168910 | 0.92 |
ENSDART00000150024
|
acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr7_-_35042805 | 0.90 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr2_+_26183544 | 0.87 |
ENSDART00000078634
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr5_-_3489302 | 0.87 |
|
|
|
| chr4_-_22950321 | 0.86 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr7_+_48727042 | 0.83 |
ENSDART00000166329
|
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr15_+_4079643 | 0.82 |
|
|
|
| chr21_-_20728623 | 0.78 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr19_-_43358940 | 0.78 |
|
|
|
| chr13_+_42280697 | 0.77 |
ENSDART00000084354
|
cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr20_-_31841557 | 0.76 |
|
|
|
| chr11_+_36827593 | 0.74 |
ENSDART00000124997
|
card19
|
caspase recruitment domain family, member 19 |
| chr4_+_13569588 | 0.74 |
ENSDART00000136152
|
calua
|
calumenin a |
| chr2_+_26647894 | 0.73 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr1_-_54294303 | 0.72 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr25_-_1390537 | 0.71 |
ENSDART00000154130
|
CR354429.1
|
ENSDARG00000097252 |
| chr19_-_46058895 | 0.70 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr5_-_19691538 | 0.69 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr7_+_48726955 | 0.68 |
ENSDART00000166329
|
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr10_+_20406765 | 0.66 |
ENSDART00000080395
|
golga7
|
golgin A7 |
| chr12_+_22448972 | 0.65 |
|
|
|
| chr3_+_36373519 | 0.64 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr5_+_32231368 | 0.60 |
ENSDART00000169358
|
ENSDARG00000095142
|
ENSDARG00000095142 |
| chr7_+_32859453 | 0.59 |
ENSDART00000142450
|
zgc:153219
|
zgc:153219 |
| chr3_+_25888520 | 0.58 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr23_-_3467855 | 0.58 |
|
|
|
| chr1_+_1964902 | 0.56 |
ENSDART00000138396
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr20_+_34497843 | 0.55 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr25_-_22821132 | 0.55 |
|
|
|
| chr6_-_2089453 | 0.53 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr23_+_3468073 | 0.52 |
|
|
|
| chr11_-_12067922 | 0.51 |
ENSDART00000110117
|
socs7
|
suppressor of cytokine signaling 7 |
| chr4_-_71730637 | 0.51 |
ENSDART00000174367
|
CABZ01032472.1
|
ENSDARG00000105801 |
| chr11_-_18305730 | 0.50 |
ENSDART00000155474
|
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| chr1_-_54294382 | 0.49 |
ENSDART00000038330
|
khsrp
|
KH-type splicing regulatory protein |
| chr10_+_6120218 | 0.49 |
ENSDART00000166799
ENSDART00000157947 |
tln1
|
talin 1 |
| chr14_-_33180554 | 0.49 |
ENSDART00000108827
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr24_-_37227441 | 0.49 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr5_+_32231323 | 0.48 |
ENSDART00000169358
|
ENSDARG00000095142
|
ENSDARG00000095142 |
| chr9_-_7419306 | 0.46 |
ENSDART00000125054
|
slc23a3
|
solute carrier family 23, member 3 |
| chr15_+_46976241 | 0.45 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr6_+_28303542 | 0.45 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_10264593 | 0.41 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr13_+_26943081 | 0.40 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| KN149679v1_-_32359 | 0.40 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr6_-_6965908 | 0.40 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr14_+_26461399 | 0.39 |
ENSDART00000125289
|
ahnak
|
AHNAK nucleoprotein |
| chr14_-_15393962 | 0.39 |
ENSDART00000161123
ENSDART00000167146 |
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr20_-_46345447 | 0.35 |
ENSDART00000100544
ENSDART00000152925 |
taar10
|
trace amine-associated receptor 10 |
| chr17_-_21180028 | 0.35 |
ENSDART00000104708
|
abhd12
|
abhydrolase domain containing 12 |
| chr5_+_52934865 | 0.35 |
ENSDART00000161682
|
BX323994.2
|
ENSDARG00000100423 |
| chr4_+_75509365 | 0.34 |
|
|
|
| chr11_-_18306222 | 0.34 |
ENSDART00000155474
|
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| chr14_-_2430754 | 0.34 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr4_+_17256021 | 0.34 |
ENSDART00000132933
|
casc1
|
cancer susceptibility candidate 1 |
| chr5_-_1296190 | 0.34 |
|
|
|
| chr17_+_31722419 | 0.33 |
ENSDART00000155073
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr11_+_44414503 | 0.32 |
|
|
|
| chr24_-_35646129 | 0.31 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr13_+_41791642 | 0.30 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr14_-_33180680 | 0.29 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr23_-_27707182 | 0.29 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr8_+_49986368 | 0.29 |
ENSDART00000156403
|
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr11_-_40416762 | 0.28 |
ENSDART00000166372
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr21_+_7807310 | 0.27 |
ENSDART00000023997
|
tbca
|
tubulin cofactor a |
| chr20_+_35352522 | 0.26 |
ENSDART00000002773
ENSDART00000137915 |
fam49a
|
family with sequence similarity 49, member A |
| chr25_+_15190474 | 0.24 |
ENSDART00000147572
ENSDART00000090330 |
hipk3a
|
homeodomain interacting protein kinase 3a |
| chr16_+_11833310 | 0.19 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr15_+_17094877 | 0.18 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr5_-_51184375 | 0.14 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr7_-_20565207 | 0.13 |
ENSDART00000052977
|
cldn15a
|
claudin 15a |
| chr2_+_56705456 | 0.11 |
|
|
|
| chr18_+_439454 | 0.11 |
ENSDART00000167841
|
ss18l2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr12_+_33818414 | 0.09 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr1_+_19156170 | 0.08 |
ENSDART00000111454
|
ENSDARG00000080019
|
ENSDARG00000080019 |
| chr7_-_32859341 | 0.04 |
ENSDART00000127006
ENSDART00000143768 |
mns1
|
meiosis-specific nuclear structural 1 |
| chr3_-_61114604 | 0.02 |
|
|
|
| chr21_+_21706999 | 0.01 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.7 | 2.0 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.5 | 3.2 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.4 | 0.9 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 1.3 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.3 | 1.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 0.9 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.3 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.0 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 1.4 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.2 | 2.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.2 | 0.7 | GO:0070178 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.2 | 2.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.2 | 2.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.1 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.1 | 1.2 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.8 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 2.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.5 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 1.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0010890 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 1.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 2.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.3 | 0.8 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.3 | 0.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.3 | 0.8 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 0.8 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 1.4 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.2 | 3.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.3 | GO:0070548 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 2.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 2.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |