Project
DANIO-CODE
Navigation
Downloads
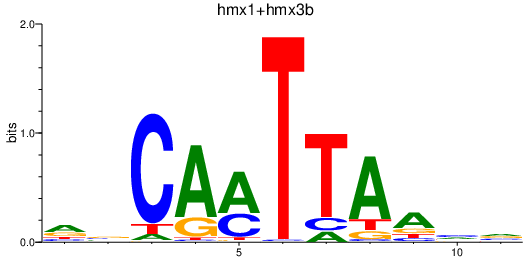
Results for hmx1+hmx3b
Z-value: 0.22
Transcription factors associated with hmx1+hmx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx1
|
ENSDARG00000095651 | H6 family homeobox 1 |
|
hmx3b
|
ENSDARG00000096716 | H6 family homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx1 | dr10_dc_chr1_-_40211790_40211795 | 0.33 | 2.2e-01 | Click! |
Activity profile of hmx1+hmx3b motif
Sorted Z-values of hmx1+hmx3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx1+hmx3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23172938 | 0.46 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr20_+_31366832 | 0.37 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr7_+_42124857 | 0.34 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr6_-_41087828 | 0.31 |
ENSDART00000028217
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr14_+_14262816 | 0.29 |
ENSDART00000164749
|
pcdh20
|
protocadherin 20 |
| chr20_+_22321303 | 0.27 |
ENSDART00000049204
|
kdr
|
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| chr21_-_39239757 | 0.25 |
|
|
|
| chr13_+_30743690 | 0.24 |
ENSDART00000134809
|
ercc6
|
excision repair cross-complementation group 6 |
| chr1_-_37990863 | 0.17 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr4_+_13932849 | 0.17 |
ENSDART00000141742
|
pphln1
|
periphilin 1 |
| chr3_+_39514598 | 0.17 |
ENSDART00000156038
|
epn2
|
epsin 2 |
| chr7_-_17129072 | 0.16 |
|
|
|
| chr12_+_2412047 | 0.16 |
ENSDART00000112032
|
ARHGAP22 (1 of many)
|
Rho GTPase activating protein 22 |
| chr6_-_37444807 | 0.15 |
ENSDART00000138351
|
cth
|
cystathionase (cystathionine gamma-lyase) |
| chr15_+_29183251 | 0.15 |
ENSDART00000153609
|
capns1b
|
calpain, small subunit 1 b |
| chr12_+_24221087 | 0.15 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr23_+_13106555 | 0.14 |
|
|
|
| chr5_-_41672394 | 0.14 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr13_+_46487890 | 0.13 |
|
|
|
| chr1_+_21037251 | 0.12 |
|
|
|
| chr4_-_26262761 | 0.11 |
ENSDART00000176623
|
BX957270.2
|
ENSDARG00000107325 |
| chr9_+_50670635 | 0.11 |
ENSDART00000179475
|
CABZ01083707.1
|
ENSDARG00000107278 |
| chr22_-_5683909 | 0.11 |
ENSDART00000138507
|
CU459089.1
|
ENSDARG00000077631 |
| chr18_+_9213713 | 0.11 |
ENSDART00000127469
ENSDART00000101192 |
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr20_-_23526954 | 0.11 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr25_-_13737344 | 0.10 |
|
|
|
| chr17_+_10437343 | 0.10 |
ENSDART00000167188
|
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr17_+_45892751 | 0.10 |
|
|
|
| chr22_+_16266827 | 0.10 |
ENSDART00000170960
|
dbt
|
dihydrolipoamide branched chain transacylase E2 |
| chr16_-_6450531 | 0.08 |
ENSDART00000060550
ENSDART00000145663 |
ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr22_-_5683814 | 0.08 |
ENSDART00000138507
|
CU459089.1
|
ENSDARG00000077631 |
| chr21_+_21870019 | 0.08 |
ENSDART00000016916
|
gria4b
|
glutamate receptor, ionotropic, AMPA 4b |
| chr20_+_28958586 | 0.08 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr25_+_17493675 | 0.08 |
ENSDART00000167750
|
vac14
|
vac14 homolog (S. cerevisiae) |
| chr17_-_52735250 | 0.08 |
|
|
|
| chr20_-_23527004 | 0.07 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_+_40292617 | 0.07 |
|
|
|
| chr4_-_70948610 | 0.07 |
ENSDART00000174299
|
CABZ01054394.2
|
ENSDARG00000098324 |
| chr1_-_37990935 | 0.07 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr23_-_28367816 | 0.06 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr3_-_3589844 | 0.06 |
ENSDART00000140482
|
ENSDARG00000079723
|
ENSDARG00000079723 |
| chr24_-_12836986 | 0.05 |
ENSDART00000133166
|
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr13_-_36785295 | 0.04 |
ENSDART00000167154
|
trim9
|
tripartite motif containing 9 |
| chr10_-_39210658 | 0.04 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr13_-_29290894 | 0.04 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr5_+_36431824 | 0.04 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr8_+_23172960 | 0.04 |
ENSDART00000028946
|
tpd52l2a
|
tumor protein D52-like 2a |
| chr4_-_18752965 | 0.04 |
ENSDART00000145747
|
atxn10
|
ataxin 10 |
| chr11_-_18081944 | 0.03 |
ENSDART00000113468
|
CABZ01112215.1
|
ENSDARG00000079534 |
| chr6_+_20266345 | 0.03 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr13_+_15084591 | 0.03 |
ENSDART00000133041
|
pank2
|
pantothenate kinase 2 |
| chr17_-_52735615 | 0.03 |
|
|
|
| chr9_+_50609926 | 0.02 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr1_-_37991246 | 0.02 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr6_+_11445617 | 0.01 |
ENSDART00000151447
ENSDART00000151618 |
calcrlb
|
calcitonin receptor-like b |
| chr10_-_39211281 | 0.01 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr19_-_6503643 | 0.01 |
|
|
|
| chr4_+_13932693 | 0.01 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr16_-_28333633 | 0.01 |
ENSDART00000121671
|
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr17_+_25828037 | 0.00 |
|
|
|
| chr9_+_50670606 | 0.00 |
ENSDART00000179475
|
CABZ01083707.1
|
ENSDARG00000107278 |
| chr19_-_35286400 | 0.00 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr5_-_66441778 | 0.00 |
ENSDART00000168245
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr18_+_17503828 | 0.00 |
ENSDART00000161827
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.2 | GO:0019344 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |