Project
DANIO-CODE
Navigation
Downloads
Results for hmx4
Z-value: 1.61
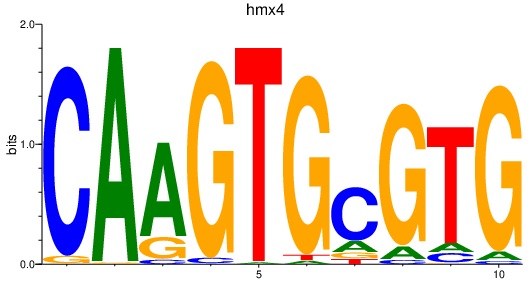
Transcription factors associated with hmx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx4
|
ENSDARG00000007941 | H6 family homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx4 | dr10_dc_chr1_-_40208469_40208513 | 0.93 | 1.4e-07 | Click! |
Activity profile of hmx4 motif
Sorted Z-values of hmx4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25157675 | 6.50 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr11_-_5868257 | 6.24 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr13_+_7575753 | 4.71 |
|
|
|
| chr6_+_17959219 | 4.32 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr20_-_26632676 | 4.17 |
ENSDART00000131994
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr5_-_7666964 | 3.77 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr1_+_25662652 | 3.69 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr13_+_22528640 | 3.63 |
ENSDART00000078877
|
sncga
|
synuclein, gamma a |
| chr1_-_24747855 | 3.56 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr13_-_33696425 | 3.52 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr22_-_27082972 | 3.36 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr11_-_25693072 | 3.25 |
ENSDART00000122011
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr20_+_30894667 | 3.24 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr15_-_41288480 | 3.20 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr16_-_7525980 | 3.17 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr6_+_59788008 | 3.14 |
|
|
|
| chr6_-_8501230 | 3.13 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr9_-_48098629 | 3.10 |
|
|
|
| chr17_+_27417635 | 3.10 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr10_+_13251463 | 3.05 |
ENSDART00000000887
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr5_-_23829861 | 3.05 |
ENSDART00000051553
|
znf703
|
zinc finger protein 703 |
| chr3_+_36282205 | 3.00 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr16_+_24032160 | 2.95 |
ENSDART00000103190
|
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr10_-_43826919 | 2.91 |
ENSDART00000039551
ENSDART00000160786 ENSDART00000099134 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr7_-_37587227 | 2.90 |
ENSDART00000173552
|
adcy7
|
adenylate cyclase 7 |
| chr9_-_53898524 | 2.81 |
ENSDART00000175619
ENSDART00000179009 |
CABZ01078244.2
|
ENSDARG00000107473 |
| chr1_-_58606982 | 2.80 |
ENSDART00000158067
|
txndc11
|
thioredoxin domain containing 11 |
| chr13_+_3533517 | 2.73 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr16_+_32605735 | 2.72 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr16_-_24602919 | 2.71 |
ENSDART00000147478
|
cadm4
|
cell adhesion molecule 4 |
| chr2_+_49837788 | 2.60 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr20_-_29517770 | 2.56 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr6_-_23193752 | 2.55 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr22_+_12327849 | 2.52 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| KN150001v1_+_14939 | 2.49 |
|
|
|
| chr2_-_21693984 | 2.49 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase-like, a |
| chr16_+_1227270 | 2.46 |
|
|
|
| chr1_+_25662910 | 2.43 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr20_-_9107294 | 2.43 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_+_27383737 | 2.39 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr10_+_7712522 | 2.37 |
ENSDART00000157608
|
fam136a
|
family with sequence similarity 136, member A |
| chr21_+_17731439 | 2.35 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| KN150702v1_+_177145 | 2.35 |
ENSDART00000166225
|
CABZ01076674.1
|
ENSDARG00000102620 |
| chr2_-_50244008 | 2.32 |
ENSDART00000029412
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr14_+_33382973 | 2.30 |
ENSDART00000132488
|
apln
|
apelin |
| chr3_+_54327353 | 2.29 |
ENSDART00000127487
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr1_+_9176411 | 2.28 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr7_-_33080261 | 2.28 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr12_-_10001043 | 2.25 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr23_-_9924987 | 2.23 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr9_-_34491458 | 2.21 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr19_+_31046291 | 2.19 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr14_-_17353336 | 2.14 |
|
|
|
| chr25_-_18234069 | 2.14 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr4_-_20435099 | 2.13 |
ENSDART00000055317
|
lrrc17
|
leucine rich repeat containing 17 |
| chr9_-_41044622 | 2.07 |
ENSDART00000143384
|
si:dkey-95p16.1
|
si:dkey-95p16.1 |
| chr7_-_64637419 | 2.07 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr1_-_22583661 | 2.05 |
ENSDART00000147800
|
rpl9
|
ribosomal protein L9 |
| chr25_+_14411153 | 2.04 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr1_-_13547500 | 2.00 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr15_-_31635737 | 2.00 |
ENSDART00000156047
|
hmgb1b
|
high mobility group box 1b |
| chr22_+_21997000 | 1.98 |
ENSDART00000046174
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr7_+_28862183 | 1.98 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr13_+_27654900 | 1.97 |
ENSDART00000160550
|
CR356236.1
|
ENSDARG00000102040 |
| chr16_+_5625301 | 1.95 |
|
|
|
| chr6_+_59762717 | 1.95 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr23_+_24711233 | 1.90 |
|
|
|
| chr21_+_5887486 | 1.88 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr23_+_6043862 | 1.88 |
|
|
|
| chr7_+_19300351 | 1.85 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr25_-_32344881 | 1.84 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr14_+_13919450 | 1.84 |
ENSDART00000174760
|
nlgn3a
|
neuroligin 3a |
| chr22_+_12746080 | 1.83 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr13_+_3533426 | 1.83 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr7_+_13753344 | 1.80 |
|
|
|
| chr4_+_5733160 | 1.79 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr19_+_7798512 | 1.79 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr13_+_51663349 | 1.77 |
|
|
|
| chr25_+_5232865 | 1.73 |
|
|
|
| chr19_+_22478256 | 1.73 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr13_+_3533318 | 1.73 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr6_+_1873957 | 1.68 |
|
|
|
| chr19_-_20614846 | 1.65 |
ENSDART00000155527
|
si:ch211-155k24.9
|
si:ch211-155k24.9 |
| chr21_-_15578808 | 1.60 |
ENSDART00000136666
|
mmp11b
|
matrix metallopeptidase 11b |
| chr1_+_27174549 | 1.59 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr13_+_25356184 | 1.56 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr16_-_20901879 | 1.54 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr8_+_53173227 | 1.54 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr25_-_13057808 | 1.54 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr16_+_26673516 | 1.54 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr11_+_18823629 | 1.53 |
|
|
|
| chr16_-_32605560 | 1.52 |
|
|
|
| chr7_-_60525749 | 1.52 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr1_-_58258428 | 1.50 |
|
|
|
| chr15_-_47696481 | 1.47 |
|
|
|
| chr3_-_19050721 | 1.46 |
ENSDART00000131503
|
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr10_-_43200499 | 1.46 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr25_+_18487408 | 1.46 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr23_+_39713307 | 1.45 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr6_-_923434 | 1.45 |
ENSDART00000167390
|
zgc:113442
|
zgc:113442 |
| chr17_+_53068731 | 1.45 |
ENSDART00000156774
|
dph6
|
diphthamine biosynthesis 6 |
| chr10_+_9134634 | 1.45 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr1_+_157674 | 1.44 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr20_-_53629996 | 1.44 |
ENSDART00000141826
|
mettl24
|
methyltransferase like 24 |
| chr16_-_33637966 | 1.42 |
ENSDART00000142965
|
BX511161.1
|
ENSDARG00000092272 |
| chr9_-_53898458 | 1.42 |
ENSDART00000175619
ENSDART00000179009 |
CABZ01078244.2
|
ENSDARG00000107473 |
| chr3_-_28078162 | 1.39 |
ENSDART00000165936
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_+_45121016 | 1.39 |
ENSDART00000172328
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr10_-_7712439 | 1.36 |
ENSDART00000159330
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr7_-_64637051 | 1.35 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr6_+_24299180 | 1.32 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| KN150207v1_-_1208 | 1.32 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr16_+_20932876 | 1.28 |
ENSDART00000079343
|
jazf1b
|
JAZF zinc finger 1b |
| chr16_+_5625552 | 1.28 |
|
|
|
| chr25_+_35782970 | 1.28 |
ENSDART00000125440
|
hist1h4l
|
histone 1, H4, like |
| chr19_+_12843381 | 1.27 |
ENSDART00000139909
|
mc5ra
|
melanocortin 5a receptor |
| chr8_-_3974357 | 1.26 |
ENSDART00000163754
ENSDART00000169474 |
mtmr3
|
myotubularin related protein 3 |
| chr1_-_18695214 | 1.25 |
|
|
|
| chr7_-_667744 | 1.24 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr14_-_33704021 | 1.22 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_-_13912235 | 1.21 |
ENSDART00000177773
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr18_-_19467100 | 1.19 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr16_+_13993746 | 1.19 |
ENSDART00000101304
|
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr20_-_53271601 | 1.18 |
|
|
|
| chr6_-_39656043 | 1.17 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr21_-_8000406 | 1.16 |
|
|
|
| chr23_-_21288972 | 1.16 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr18_-_36085063 | 1.16 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr3_+_55779906 | 1.16 |
|
|
|
| chr1_-_54294303 | 1.16 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr14_-_33483075 | 1.16 |
ENSDART00000158870
|
si:dkey-76i15.1
|
si:dkey-76i15.1 |
| chr23_+_200650 | 1.14 |
|
|
|
| chr8_+_52633069 | 1.14 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr4_-_9591451 | 1.14 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_+_23093475 | 1.11 |
|
|
|
| chr17_+_27706409 | 1.10 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr17_+_48349119 | 1.09 |
ENSDART00000019804
|
CABZ01043017.1
|
ENSDARG00000001937 |
| chr7_+_25587183 | 1.09 |
ENSDART00000148780
|
mtmr1a
|
myotubularin related protein 1a |
| chr17_-_26849495 | 1.09 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr9_-_41982635 | 1.09 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr3_+_32234028 | 1.09 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr7_-_57030751 | 1.06 |
|
|
|
| chr24_-_42226496 | 1.05 |
ENSDART00000163208
ENSDART00000112680 |
rmdn1
|
regulator of microtubule dynamics 1 |
| chr13_-_36420228 | 1.04 |
|
|
|
| chr14_+_33382672 | 1.04 |
ENSDART00000075312
|
apln
|
apelin |
| chr3_+_56507117 | 1.03 |
ENSDART00000154405
|
rac1b
|
ras-related C3 botulinum toxin substrate 1b (rho family, small GTP binding protein Rac1) |
| chr3_+_59491893 | 1.03 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr10_-_38206238 | 1.02 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr15_+_42393715 | 1.01 |
|
|
|
| chr14_-_24744825 | 0.98 |
ENSDART00000159827
ENSDART00000131027 |
TMEM216
|
transmembrane protein 216 |
| chr23_+_4385722 | 0.98 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr22_-_12130441 | 0.96 |
ENSDART00000146785
|
tmem163b
|
transmembrane protein 163b |
| chr6_+_39838929 | 0.94 |
ENSDART00000112637
|
smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr23_-_26562635 | 0.94 |
ENSDART00000168052
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr1_+_35462641 | 0.94 |
ENSDART00000053773
ENSDART00000147458 |
lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr24_-_39132141 | 0.93 |
|
|
|
| chr13_-_49546922 | 0.93 |
|
|
|
| chr20_+_38129564 | 0.93 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr20_-_53630053 | 0.91 |
ENSDART00000135091
|
mettl24
|
methyltransferase like 24 |
| chr23_+_22408855 | 0.90 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr8_-_24788837 | 0.89 |
ENSDART00000126220
|
alx3
|
ALX homeobox 3 |
| chr18_+_37034218 | 0.88 |
|
|
|
| chr3_-_22699157 | 0.88 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr3_+_16116080 | 0.87 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr21_-_19789339 | 0.86 |
|
|
|
| chr7_-_42972804 | 0.86 |
ENSDART00000054552
|
cdh8
|
cadherin 8 |
| chr10_+_13251650 | 0.85 |
ENSDART00000136932
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr14_-_46648350 | 0.84 |
|
|
|
| chr7_+_22272746 | 0.84 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr17_-_50343403 | 0.84 |
ENSDART00000140121
|
si:ch211-235i11.5
|
si:ch211-235i11.5 |
| chr13_-_24248675 | 0.83 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr1_+_45148094 | 0.83 |
ENSDART00000048191
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr16_-_28775103 | 0.83 |
ENSDART00000127753
|
adam15
|
ADAM metallopeptidase domain 15 |
| chr20_-_32429358 | 0.81 |
ENSDART00000062982
|
foxo3b
|
forkhead box O3b |
| KN150001v1_+_15017 | 0.81 |
|
|
|
| chr10_-_20681689 | 0.79 |
|
|
|
| chr6_-_39655998 | 0.79 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr6_-_23193873 | 0.79 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr6_-_34954414 | 0.78 |
|
|
|
| chr25_+_9904821 | 0.78 |
|
|
|
| chr17_+_31604804 | 0.76 |
ENSDART00000111629
|
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr16_+_4825665 | 0.73 |
|
|
|
| chr13_+_7575720 | 0.73 |
|
|
|
| chr23_-_26562855 | 0.72 |
ENSDART00000168052
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr10_-_175160 | 0.72 |
|
|
|
| chr16_+_54729975 | 0.71 |
|
|
|
| chr13_+_41791642 | 0.71 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr6_+_46404650 | 0.70 |
ENSDART00000131203
ENSDART00000103472 ENSDART00000132845 ENSDART00000138567 ENSDART00000168440 |
pbrm1l
|
polybromo 1, like |
| chr11_-_2227358 | 0.69 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr24_-_16878970 | 0.69 |
ENSDART00000106058
ENSDART00000106057 |
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr4_-_4583387 | 0.69 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr13_+_18401965 | 0.68 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr23_+_17220363 | 0.67 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr12_-_26323696 | 0.67 |
|
|
|
| chr24_-_20176227 | 0.67 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr16_-_54638972 | 0.66 |
|
|
|
| chr17_+_25314291 | 0.66 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr11_+_40567461 | 0.66 |
ENSDART00000160023
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr3_-_33290990 | 0.65 |
ENSDART00000075493
ENSDART00000132293 |
ccdc103
|
coiled-coil domain containing 103 |
| chr11_-_18656380 | 0.65 |
ENSDART00000080355
|
ENSDARG00000057708
|
ENSDARG00000057708 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.2 | 6.1 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 1.1 | 3.2 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.9 | 2.7 | GO:0090171 | chondrocyte morphogenesis(GO:0090171) |
| 0.9 | 2.7 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.8 | 3.4 | GO:0097535 | trigeminal sensory nucleus development(GO:0021730) lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.8 | 2.4 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.8 | 2.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.7 | 2.1 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.6 | 1.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.5 | 2.9 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 4.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.5 | 1.8 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.5 | 1.4 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.5 | 2.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.4 | 1.6 | GO:0010657 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.4 | 5.0 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.3 | 3.0 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.3 | 2.9 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.3 | 3.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.3 | 2.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 3.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 1.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 2.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.8 | GO:0050687 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.2 | 2.1 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 2.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 0.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 3.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 2.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 4.3 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 2.3 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.9 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 1.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 1.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 2.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 1.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 1.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 2.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 2.7 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 0.3 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.7 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.3 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 1.6 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 6.5 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 7.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 1.0 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.1 | 0.3 | GO:0060359 | sperm motility(GO:0030317) response to alkaloid(GO:0043279) response to ammonium ion(GO:0060359) cellular response to ammonium ion(GO:0071242) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.6 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 2.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 2.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.0 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 1.3 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.4 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 3.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.1 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 1.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 2.9 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 2.3 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 1.5 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 2.5 | GO:0042127 | regulation of cell proliferation(GO:0042127) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 1.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 1.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 2.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 3.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 1.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 0.7 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 2.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 3.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.2 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 5.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 4.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 17.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 3.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 3.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 1.2 | 6.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.2 | 3.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.9 | 4.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.8 | 2.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 2.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.6 | 1.8 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.6 | 6.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.5 | 3.2 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.5 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.4 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.4 | 2.6 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 3.4 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.4 | 1.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 3.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 9.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 2.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 0.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 2.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 2.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.9 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 2.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0051192 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 5.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 2.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 3.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 3.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.1 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 1.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 2.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 2.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.5 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.1 | 0.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 3.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |