Project
DANIO-CODE
Navigation
Downloads
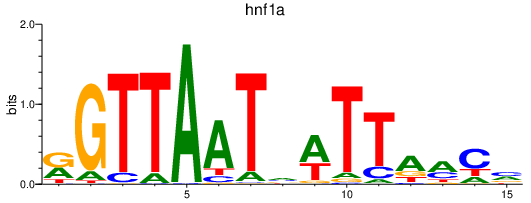
Results for hnf1a
Z-value: 2.70
Transcription factors associated with hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1a
|
ENSDARG00000009470 | HNF1 homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1a | dr10_dc_chr8_+_39768744_39768780 | 0.93 | 1.3e-07 | Click! |
Activity profile of hnf1a motif
Sorted Z-values of hnf1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_99353 | 14.15 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr8_+_933677 | 11.62 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr22_-_17570306 | 11.13 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr24_-_39722595 | 10.46 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr2_-_18157106 | 10.00 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr12_-_6144233 | 9.47 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr22_-_23598571 | 9.16 |
ENSDART00000166800
|
cfhl4
|
complement factor H like 4 |
| chr1_+_9367672 | 8.61 |
ENSDART00000144756
|
fgb
|
fibrinogen beta chain |
| chr21_-_23271127 | 8.47 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr16_-_42990753 | 8.10 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr1_-_11439671 | 8.03 |
ENSDART00000164817
|
mtp
|
microsomal triglyceride transfer protein |
| chr20_+_18841440 | 7.67 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr7_-_25623974 | 7.58 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr19_-_22802777 | 7.56 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr14_-_46651359 | 7.10 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr5_+_31745031 | 6.97 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr14_-_20760787 | 6.84 |
|
|
|
| chr14_-_31128891 | 6.51 |
|
|
|
| chr25_+_30746445 | 6.49 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr6_+_7742042 | 6.42 |
ENSDART00000112290
ENSDART00000062740 |
palm3
|
paralemmin 3 |
| chr21_+_25199691 | 6.41 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr3_-_16100715 | 6.18 |
ENSDART00000146699
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr7_+_26978970 | 6.14 |
ENSDART00000150068
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr17_+_3879416 | 5.88 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr12_+_31558667 | 5.72 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr13_-_23536022 | 5.46 |
|
|
|
| chr9_-_43178744 | 5.43 |
|
|
|
| chr15_-_24987099 | 5.37 |
|
|
|
| chr23_+_32409339 | 5.10 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr13_-_40600924 | 5.07 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr16_-_9785057 | 5.03 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr21_-_44610563 | 5.01 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr18_+_9413588 | 5.00 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr3_+_15355472 | 4.98 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr11_-_23079615 | 4.98 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr3_+_45375639 | 4.86 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr15_-_16075172 | 4.85 |
|
|
|
| chr15_+_27431469 | 4.60 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr22_+_3136570 | 4.52 |
ENSDART00000163327
|
rpl36
|
ribosomal protein L36 |
| chr23_+_24711233 | 4.42 |
|
|
|
| chr4_-_885413 | 4.40 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr13_-_11404389 | 4.40 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr9_+_21340251 | 4.35 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr5_-_55843553 | 4.30 |
ENSDART00000024207
|
tbx2a
|
T-box 2a |
| chr16_+_55351732 | 4.23 |
ENSDART00000109391
|
CABZ01052290.1
|
ENSDARG00000077559 |
| chr20_-_7303735 | 4.03 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr10_+_7296895 | 4.03 |
ENSDART00000161506
|
fut10
|
fucosyltransferase 10 |
| chr10_-_8074713 | 4.00 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase, H+ transporting, lysosomal V0 subunit a2a |
| chr8_-_47811709 | 4.00 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr24_-_16895179 | 3.90 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr13_-_42453219 | 3.78 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| chr6_+_30469823 | 3.71 |
ENSDART00000121492
|
FP236735.1
|
ENSDARG00000087805 |
| chr22_+_12746080 | 3.67 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr14_-_46650724 | 3.64 |
|
|
|
| chr20_-_7303804 | 3.62 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr5_-_62747812 | 3.54 |
|
|
|
| chr6_-_39346614 | 3.52 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr22_-_23641813 | 3.39 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr14_+_8198019 | 3.34 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr24_+_9124068 | 3.32 |
|
|
|
| chr1_-_22160662 | 3.30 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr6_+_7741987 | 3.20 |
ENSDART00000112290
ENSDART00000062740 |
palm3
|
paralemmin 3 |
| chr17_-_13072982 | 3.15 |
ENSDART00000178859
|
CABZ01047998.1
|
ENSDARG00000105945 |
| chr12_+_15624640 | 3.13 |
ENSDART00000079803
|
nmt1b
|
N-myristoyltransferase 1b |
| chr5_-_54151518 | 3.07 |
|
|
|
| chr11_-_40415291 | 2.97 |
|
|
|
| chr20_-_25582221 | 2.95 |
ENSDART00000157559
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr17_-_35004687 | 2.85 |
|
|
|
| chr2_+_49683715 | 2.80 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr15_-_27431216 | 2.80 |
ENSDART00000155933
|
AL954322.2
|
ENSDARG00000097072 |
| chr22_+_34738389 | 2.78 |
ENSDART00000154372
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr23_+_31505372 | 2.76 |
ENSDART00000138350
|
rps12
|
ribosomal protein S12 |
| chr5_+_3721680 | 2.70 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr8_+_1122000 | 2.67 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr5_+_44722544 | 2.65 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr1_-_18809429 | 2.61 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr3_-_34208423 | 2.57 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr15_+_9321225 | 2.56 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr5_-_62747914 | 2.54 |
|
|
|
| chr3_+_54506912 | 2.53 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr9_-_1457681 | 2.53 |
ENSDART00000124777
ENSDART00000093412 |
osbpl6
|
oxysterol binding protein-like 6 |
| chr25_-_7346766 | 2.50 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr21_-_99497 | 2.46 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr11_-_37730181 | 2.44 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr11_-_27455348 | 2.43 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr3_+_45375698 | 2.43 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr24_+_19842162 | 2.36 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr21_+_43674754 | 2.35 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr7_+_39934376 | 2.35 |
ENSDART00000173742
|
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr6_+_54213569 | 2.31 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr14_+_721560 | 2.26 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr17_+_15026344 | 2.19 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr8_+_26502909 | 2.17 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr7_-_25624212 | 2.17 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr5_-_30114979 | 2.16 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr9_-_43736549 | 2.13 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr8_+_13069277 | 2.08 |
ENSDART00000138433
|
itgb4
|
integrin, beta 4 |
| chr17_+_15025997 | 2.04 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr3_+_23607148 | 2.03 |
ENSDART00000143981
|
hoxb3a
|
homeobox B3a |
| chr5_-_9162684 | 2.01 |
|
|
|
| chr9_-_14137664 | 2.00 |
ENSDART00000132747
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr3_+_15355424 | 1.99 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr21_-_44109455 | 1.95 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr10_-_41742329 | 1.95 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr3_+_40028301 | 1.95 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr10_-_1799784 | 1.92 |
ENSDART00000058627
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr15_-_23532475 | 1.85 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr2_-_44893769 | 1.84 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr9_+_33177527 | 1.81 |
ENSDART00000174533
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr17_-_14956738 | 1.78 |
|
|
|
| chr9_-_48152388 | 1.73 |
|
|
|
| chr7_-_25624128 | 1.68 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr10_+_4476558 | 1.68 |
ENSDART00000125299
|
plk2a
|
polo-like kinase 2a (Drosophila) |
| chr5_-_37285874 | 1.67 |
ENSDART00000132152
|
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr5_+_8843643 | 1.66 |
ENSDART00000149417
ENSDART00000061616 |
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr13_+_48327287 | 1.65 |
ENSDART00000168596
|
si:ch1073-268j14.1
|
si:ch1073-268j14.1 |
| chr2_-_24661477 | 1.61 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr14_-_32543646 | 1.60 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr16_-_8711133 | 1.60 |
|
|
|
| chr1_+_25974174 | 1.59 |
ENSDART00000152144
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr14_-_46650551 | 1.59 |
|
|
|
| chr24_-_8592157 | 1.58 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr16_+_11833310 | 1.51 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr20_-_7141247 | 1.46 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr2_-_5492583 | 1.46 |
ENSDART00000152907
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr17_+_15026176 | 1.43 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr24_+_23646663 | 1.43 |
ENSDART00000146580
|
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr4_-_30124515 | 1.38 |
|
|
|
| chr14_-_46650640 | 1.27 |
|
|
|
| chr1_+_25974337 | 1.26 |
ENSDART00000152617
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr23_+_25930072 | 1.26 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| KN149681v1_-_46883 | 1.23 |
|
|
|
| chr9_+_30909291 | 1.22 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr20_-_25537241 | 1.20 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr9_+_30183691 | 1.16 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr16_+_26573825 | 1.16 |
|
|
|
| chr9_+_30909382 | 1.13 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr10_+_20151165 | 1.13 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr4_+_74835204 | 1.06 |
ENSDART00000172647
ENSDART00000174351 |
zgc:113209
|
zgc:113209 |
| chr1_+_25974264 | 1.04 |
ENSDART00000152785
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr5_+_67157870 | 1.00 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr22_-_17570275 | 1.00 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr18_+_9413668 | 0.99 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr22_-_23721742 | 0.94 |
ENSDART00000163228
|
cfh
|
complement factor H |
| chr24_+_14569372 | 0.93 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr6_+_32478052 | 0.90 |
|
|
|
| chr18_+_10957421 | 0.86 |
ENSDART00000166561
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr3_+_26603352 | 0.86 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr3_+_52645074 | 0.85 |
ENSDART00000058958
|
atp13a1
|
ATPase type 13A1 |
| chr12_-_20251183 | 0.82 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr20_-_7303680 | 0.82 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr19_+_20690151 | 0.73 |
ENSDART00000151059
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr5_-_54151599 | 0.72 |
|
|
|
| chr17_+_11950934 | 0.70 |
|
|
|
| chr4_-_70791859 | 0.69 |
ENSDART00000125726
|
si:ch211-161m3.6
|
si:ch211-161m3.6 |
| chr11_-_27832530 | 0.66 |
ENSDART00000172937
|
BX248318.1
|
ENSDARG00000104418 |
| chr19_-_15324958 | 0.64 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr3_-_31583910 | 0.63 |
ENSDART00000051542
|
ccdc47
|
coiled-coil domain containing 47 |
| chr7_-_10318692 | 0.60 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr8_+_13069233 | 0.56 |
ENSDART00000138433
|
itgb4
|
integrin, beta 4 |
| chr7_-_61591422 | 0.55 |
ENSDART00000110328
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr21_+_39055276 | 0.50 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr8_+_13069201 | 0.50 |
ENSDART00000138433
|
itgb4
|
integrin, beta 4 |
| chr11_-_6071086 | 0.48 |
ENSDART00000122262
|
babam1
|
BRISC and BRCA1 A complex member 1 |
| chr5_-_28888704 | 0.47 |
|
|
|
| chr12_+_20545413 | 0.46 |
ENSDART00000144804
|
mxra7
|
matrix-remodelling associated 7 |
| chr3_+_30967192 | 0.41 |
ENSDART00000153354
|
BX004999.1
|
ENSDARG00000096792 |
| chr7_+_41033650 | 0.40 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr11_+_6138968 | 0.40 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr14_-_46650934 | 0.40 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr10_+_2948774 | 0.35 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr5_-_28888658 | 0.33 |
|
|
|
| chr9_+_28877812 | 0.33 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr18_+_5005303 | 0.32 |
|
|
|
| chr25_-_36621405 | 0.27 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr10_+_20151082 | 0.27 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr21_-_99613 | 0.14 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr2_-_27953540 | 0.14 |
ENSDART00000040555
|
tgs1
|
trimethylguanosine synthase 1 |
| chr9_-_30692456 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.7 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 3.3 | 10.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 3.0 | 8.9 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 2.3 | 6.8 | GO:1902855 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 2.1 | 8.5 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 1.5 | 5.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.4 | 7.0 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 1.3 | 7.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.2 | 3.7 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 1.2 | 6.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 1.1 | 8.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 6.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.9 | 8.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.8 | 5.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.8 | 2.4 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.8 | 3.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.7 | 5.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.7 | 9.5 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.6 | 4.0 | GO:0036065 | fucosylation(GO:0036065) |
| 0.6 | 3.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.5 | 5.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.5 | 1.9 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.5 | 2.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.4 | 2.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.4 | 1.6 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) purine-containing compound transmembrane transport(GO:0072530) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.4 | 2.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 2.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.4 | 4.0 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.3 | 2.3 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.3 | 2.5 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 1.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 6.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 3.8 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 2.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 14.2 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 11.0 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 0.5 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 1.6 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.1 | 0.4 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 2.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 4.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 4.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.4 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 1.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 2.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 1.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 14.1 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 2.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.8 | GO:0072001 | kidney development(GO:0001822) renal system development(GO:0072001) |
| 0.0 | 1.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.4 | 7.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 14.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 4.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 2.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 4.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 0.5 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 4.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 6.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 5.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 2.8 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 2.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 8.5 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 2.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 13.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 10.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 16.7 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 4.0 | 12.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 2.3 | 11.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 1.7 | 5.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.3 | 4.0 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.2 | 3.7 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 1.1 | 11.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.9 | 2.7 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.8 | 3.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.7 | 10.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.6 | 1.9 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.6 | 8.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 3.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.5 | 5.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 2.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.4 | 1.6 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.4 | 4.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.3 | 2.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 5.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.3 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 5.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 2.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 0.8 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 2.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 9.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 10.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 1.8 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 2.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 8.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 2.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 5.1 | GO:0031406 | carboxylic acid binding(GO:0031406) |
| 0.1 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 2.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 7.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 20.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.3 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 2.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 6.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 2.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.4 | 8.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.2 | 10.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 16.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 7.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 8.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 4.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 10.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 2.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 2.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 7.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 8.5 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 5.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |