Project
DANIO-CODE
Navigation
Downloads
Results for hnf1ba+hnf1bb
Z-value: 2.65
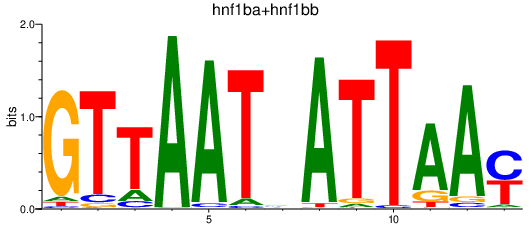
Transcription factors associated with hnf1ba+hnf1bb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1ba
|
ENSDARG00000006615 | HNF1 homeobox Ba |
|
hnf1bb
|
ENSDARG00000022295 | HNF1 homeobox Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1bb | dr10_dc_chr21_+_38770125_38770137 | 0.92 | 4.3e-07 | Click! |
| hnf1ba | dr10_dc_chr15_-_16076874_16076952 | 0.79 | 2.8e-04 | Click! |
Activity profile of hnf1ba+hnf1bb motif
Sorted Z-values of hnf1ba+hnf1bb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1ba+hnf1bb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_25623974 | 11.53 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr2_-_18157106 | 10.58 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr16_-_42990753 | 10.05 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr21_-_23271127 | 9.60 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr19_-_5435448 | 9.21 |
ENSDART00000027701
|
krt92
|
keratin 92 |
| chr1_-_11439671 | 8.73 |
ENSDART00000164817
|
mtp
|
microsomal triglyceride transfer protein |
| chr12_-_6144233 | 8.65 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr1_+_9367672 | 7.66 |
ENSDART00000144756
|
fgb
|
fibrinogen beta chain |
| chr24_-_39722595 | 7.23 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr21_-_20291707 | 7.17 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_+_31745031 | 7.14 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr14_-_20760787 | 7.08 |
|
|
|
| chr15_-_18159904 | 7.02 |
ENSDART00000170874
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr6_+_7742042 | 6.74 |
ENSDART00000112290
ENSDART00000062740 |
palm3
|
paralemmin 3 |
| chr3_-_16100715 | 6.61 |
ENSDART00000146699
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr15_+_35184996 | 6.51 |
ENSDART00000086954
ENSDART00000161594 |
sesn3
|
sestrin 3 |
| chr14_-_31128891 | 6.37 |
|
|
|
| chr8_-_985673 | 6.26 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr13_-_23536022 | 6.17 |
|
|
|
| chr19_+_20199890 | 5.44 |
ENSDART00000161019
|
hoxa4a
|
homeobox A4a |
| chr10_+_25407847 | 5.38 |
ENSDART00000047541
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr16_-_9785057 | 5.15 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr5_-_40707316 | 4.97 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr8_-_53508979 | 4.93 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr13_-_18704185 | 4.85 |
ENSDART00000146795
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr24_-_6048914 | 4.70 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr14_-_46651359 | 4.65 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr9_+_38814714 | 4.60 |
ENSDART00000114728
|
znf148
|
zinc finger protein 148 |
| chr9_+_21340251 | 4.44 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr2_+_2374651 | 4.26 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr23_+_32409339 | 4.24 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr8_-_47811709 | 4.19 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr24_+_35899507 | 4.14 |
ENSDART00000122408
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr20_-_35938683 | 4.03 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr22_+_20436009 | 4.00 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr6_+_30469823 | 3.99 |
ENSDART00000121492
|
FP236735.1
|
ENSDARG00000087805 |
| chr20_-_25582221 | 3.96 |
ENSDART00000157559
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr5_-_11562856 | 3.94 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr17_-_13072982 | 3.85 |
ENSDART00000178859
|
CABZ01047998.1
|
ENSDARG00000105945 |
| chr5_+_44722544 | 3.82 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr23_+_24557956 | 3.72 |
|
|
|
| chr7_-_25624212 | 3.71 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr6_+_7741987 | 3.59 |
ENSDART00000112290
ENSDART00000062740 |
palm3
|
paralemmin 3 |
| chr6_+_60060297 | 3.53 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr23_+_24711233 | 3.50 |
|
|
|
| chr1_-_22160662 | 3.49 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr15_+_28269289 | 3.47 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr21_+_43674754 | 3.46 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr22_-_23641813 | 3.38 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr24_-_16895179 | 3.29 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr15_+_42626315 | 3.25 |
|
|
|
| chr14_+_38313985 | 3.22 |
ENSDART00000148938
|
rnasel3
|
ribonuclease like 3 |
| chr25_+_23326562 | 3.14 |
|
|
|
| chr1_-_18809429 | 3.13 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr5_+_3721680 | 3.11 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr17_+_15026344 | 3.07 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr7_-_25624128 | 3.05 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr8_+_15216833 | 3.02 |
ENSDART00000141185
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr2_-_24661477 | 3.01 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr17_+_15025997 | 3.01 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr11_-_37730181 | 2.96 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr13_-_40600924 | 2.91 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr6_+_54213569 | 2.89 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr24_+_25767893 | 2.83 |
ENSDART00000143099
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr14_+_721560 | 2.82 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr5_-_54151518 | 2.73 |
|
|
|
| chr1_-_58258428 | 2.71 |
|
|
|
| chr5_+_37254520 | 2.64 |
ENSDART00000051222
|
ins
|
preproinsulin |
| chr7_-_72027936 | 2.55 |
ENSDART00000161914
|
slc35c1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr21_+_39918070 | 2.39 |
ENSDART00000135235
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr12_-_3903961 | 2.25 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr24_+_9124068 | 2.22 |
|
|
|
| chr3_+_40028301 | 2.17 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr25_+_23326630 | 2.15 |
|
|
|
| chr17_+_15026176 | 2.11 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr4_-_885413 | 2.11 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr5_-_30114979 | 2.09 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr7_+_26788074 | 2.07 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr11_-_44738307 | 2.04 |
ENSDART00000167540
|
afmid
|
arylformamidase |
| chr22_+_34738389 | 2.01 |
ENSDART00000154372
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr24_+_19842162 | 1.88 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr9_-_14137664 | 1.87 |
ENSDART00000132747
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr4_+_74835204 | 1.84 |
ENSDART00000172647
ENSDART00000174351 |
zgc:113209
|
zgc:113209 |
| chr23_+_25930072 | 1.79 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr18_-_48498261 | 1.78 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr9_+_30909291 | 1.74 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr8_-_19166630 | 1.73 |
|
|
|
| chr11_-_27832530 | 1.68 |
ENSDART00000172937
|
BX248318.1
|
ENSDARG00000104418 |
| chr13_-_36409205 | 1.65 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr16_-_8711133 | 1.63 |
|
|
|
| chr23_+_28843683 | 1.51 |
ENSDART00000132179
|
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr9_+_30183691 | 1.50 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr24_+_23646663 | 1.47 |
ENSDART00000146580
|
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr9_+_30909382 | 1.46 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr5_-_42304154 | 1.37 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr16_-_22903823 | 1.32 |
|
|
|
| chr3_+_26603352 | 1.30 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr10_-_41742329 | 1.28 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr1_+_25974174 | 1.18 |
ENSDART00000152144
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr6_-_43030291 | 1.17 |
ENSDART00000149637
|
glyctk
|
glycerate kinase |
| chr8_+_26502909 | 1.17 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr2_+_51087570 | 1.11 |
|
|
|
| chr5_-_37285874 | 1.09 |
ENSDART00000132152
|
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr24_-_21768269 | 1.05 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr19_-_15324958 | 1.03 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr8_+_30690195 | 1.02 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr5_+_23625065 | 0.98 |
ENSDART00000029719
|
si:ch211-114c12.2
|
si:ch211-114c12.2 |
| chr6_+_32478052 | 0.97 |
|
|
|
| chr1_+_25974337 | 0.96 |
ENSDART00000152617
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr25_+_3221696 | 0.95 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr24_+_14569372 | 0.93 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr14_-_46650551 | 0.90 |
|
|
|
| chr22_-_23721742 | 0.89 |
ENSDART00000163228
|
cfh
|
complement factor H |
| chr3_+_52645074 | 0.84 |
ENSDART00000058958
|
atp13a1
|
ATPase type 13A1 |
| chr17_+_11950934 | 0.79 |
|
|
|
| chr15_+_44442345 | 0.79 |
ENSDART00000028011
|
msantd4
|
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr1_+_25974264 | 0.79 |
ENSDART00000152785
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr24_+_25767966 | 0.77 |
ENSDART00000143099
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr7_-_10318692 | 0.76 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr25_-_36621405 | 0.74 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr7_-_72027580 | 0.73 |
ENSDART00000161914
|
slc35c1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr14_-_46650640 | 0.71 |
|
|
|
| chr25_-_3633631 | 0.67 |
ENSDART00000159335
ENSDART00000088077 |
zgc:158398
|
zgc:158398 |
| chr22_+_10630419 | 0.65 |
ENSDART00000105835
|
tusc2b
|
tumor suppressor candidate 2b |
| chr17_+_5740225 | 0.63 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr17_-_22047445 | 0.57 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr21_+_39055276 | 0.56 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr3_+_55994886 | 0.56 |
|
|
|
| chr5_-_54151599 | 0.53 |
|
|
|
| chr20_+_34814623 | 0.49 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr7_-_32562435 | 0.49 |
ENSDART00000099872
ENSDART00000099871 ENSDART00000147554 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr5_-_11562934 | 0.46 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr24_-_21740552 | 0.42 |
|
|
|
| chr20_+_31173383 | 0.40 |
ENSDART00000136255
|
otofa
|
otoferlin a |
| chr17_-_17928974 | 0.38 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr15_+_38319701 | 0.35 |
ENSDART00000122134
|
stim1a
|
stromal interaction molecule 1a |
| chr10_+_2948774 | 0.33 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr2_+_51087643 | 0.32 |
|
|
|
| chr17_-_51804002 | 0.18 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr9_+_28877812 | 0.17 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr17_+_15025962 | 0.13 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr19_+_8225252 | 0.11 |
ENSDART00000147218
|
efna3a
|
ephrin-A3a |
| chr12_-_3904030 | 0.09 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr10_-_42302932 | 0.09 |
ENSDART00000076693
ENSDART00000073631 |
stambpa
|
STAM binding protein a |
| chr14_-_46650934 | 0.08 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr2_-_27953540 | 0.03 |
ENSDART00000040555
|
tgs1
|
trimethylguanosine synthase 1 |
| chr21_-_26678542 | 0.02 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 2.4 | 9.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 2.3 | 11.4 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 1.3 | 4.0 | GO:0050864 | regulation of B cell activation(GO:0050864) |
| 1.2 | 8.4 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 1.0 | 3.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.9 | 3.5 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.9 | 5.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.8 | 5.9 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.8 | 7.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.7 | 2.6 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of feeding behavior(GO:0060259) |
| 0.6 | 5.1 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.6 | 4.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.6 | 4.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.6 | 8.6 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.5 | 3.3 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.5 | 3.0 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.4 | 3.1 | GO:0038093 | B cell differentiation(GO:0030183) Fc receptor signaling pathway(GO:0038093) |
| 0.4 | 2.9 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.4 | 3.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.4 | 3.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 2.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.3 | 8.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.3 | 1.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 1.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.3 | 5.0 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.2 | 1.4 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.2 | 0.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.6 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 3.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.8 | GO:0035999 | folic acid-containing compound biosynthetic process(GO:0009396) tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 3.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 21.0 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 3.8 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 5.4 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 1.1 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 3.5 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 2.9 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 4.6 | GO:0030097 | hemopoiesis(GO:0030097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.3 | 11.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.6 | 5.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 5.9 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 7.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 9.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 4.9 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 8.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 5.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 9.7 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 15.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 17.2 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 8.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 33.3 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.9 | 3.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.8 | 3.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 5.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 1.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 5.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 2.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.4 | 3.0 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.4 | 1.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 8.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.4 | 2.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.4 | 5.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 2.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 4.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 3.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 0.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 0.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 0.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 4.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 4.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.2 | 10.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 8.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 1.4 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.2 | 3.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 4.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 6.6 | GO:0031406 | carboxylic acid binding(GO:0031406) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 3.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 5.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.5 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 3.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 3.3 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 9.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 20.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 8.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 20.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 1.6 | 4.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 1.5 | 7.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 7.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 2.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 6.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 4.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 2.8 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.2 | 3.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 7.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 8.6 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 4.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |