Project
DANIO-CODE
Navigation
Downloads
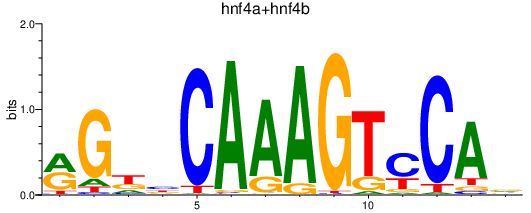
Results for hnf4a+hnf4b
Z-value: 1.18
Transcription factors associated with hnf4a+hnf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf4a
|
ENSDARG00000021494 | hepatocyte nuclear factor 4, alpha |
|
hnf4b
|
ENSDARG00000104742 | hepatic nuclear factor 4, beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf4a | dr10_dc_chr23_+_25930072_25930158 | 0.58 | 1.9e-02 | Click! |
Activity profile of hnf4a+hnf4b motif
Sorted Z-values of hnf4a+hnf4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf4a+hnf4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23999029 | 5.19 |
ENSDART00000129525
|
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr3_-_31672763 | 3.83 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr14_+_20809272 | 3.24 |
ENSDART00000139865
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr25_+_21732255 | 3.22 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr22_+_37934447 | 3.12 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr6_-_39315024 | 3.09 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr25_+_5162045 | 3.01 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr2_+_24881103 | 2.90 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr21_-_37509454 | 2.86 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr16_+_55167489 | 2.86 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr16_+_24032160 | 2.84 |
ENSDART00000103190
|
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr14_+_6122948 | 2.80 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr2_-_10667156 | 2.69 |
ENSDART00000081196
|
scinlb
|
scinderin like b |
| chr12_-_5085227 | 2.68 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr24_-_33817169 | 2.64 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr2_+_30932612 | 2.59 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr12_+_27032862 | 2.56 |
|
|
|
| chr7_+_23843682 | 2.49 |
ENSDART00000173899
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr4_-_16461748 | 2.43 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr21_+_25199691 | 2.28 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr2_+_3701942 | 2.27 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr20_+_2255204 | 2.26 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr2_+_32036450 | 2.12 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr17_+_1058137 | 2.11 |
|
|
|
| chr12_+_29994735 | 2.09 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr2_-_20941256 | 2.07 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr1_-_18695214 | 2.05 |
|
|
|
| chr5_+_51392394 | 2.04 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr23_-_15442032 | 2.01 |
ENSDART00000082060
|
ENSDARG00000078145
|
ENSDARG00000078145 |
| chr10_-_6453139 | 2.01 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr13_+_16130212 | 1.95 |
ENSDART00000125813
|
mocos
|
molybdenum cofactor sulfurase |
| chr15_-_26619404 | 1.91 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr17_-_30685367 | 1.83 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr20_+_23643050 | 1.80 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr1_-_46015367 | 1.79 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr7_-_54407681 | 1.78 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr11_+_30073930 | 1.76 |
ENSDART00000127075
|
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr4_+_11174186 | 1.73 |
ENSDART00000138661
|
tspan11
|
tetraspanin 11 |
| chr5_+_8415025 | 1.71 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr22_-_20101177 | 1.71 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| KN149726v1_+_1094 | 1.68 |
|
|
|
| chr16_+_17061069 | 1.68 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr7_+_25649559 | 1.68 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr11_+_30048913 | 1.66 |
|
|
|
| chr8_+_7740132 | 1.63 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr7_-_64637419 | 1.62 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr1_+_52739724 | 1.59 |
ENSDART00000159220
|
ucp1
|
uncoupling protein 1 |
| chr7_+_31567166 | 1.55 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr12_-_31342432 | 1.54 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr2_-_37974443 | 1.54 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr13_+_28689749 | 1.51 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr11_+_8119829 | 1.50 |
ENSDART00000011183
|
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr3_-_60973097 | 1.47 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr17_+_443558 | 1.45 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr19_+_31944725 | 1.44 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr3_-_10342940 | 1.43 |
ENSDART00000124419
|
BX539325.2
|
ENSDARG00000101154 |
| chr24_-_33817036 | 1.31 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr22_+_16293900 | 1.29 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr12_-_27032151 | 1.27 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr8_+_1760968 | 1.27 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr23_+_17853977 | 1.26 |
|
|
|
| chr15_-_1225595 | 1.23 |
|
|
|
| chr3_-_23513177 | 1.22 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr22_+_16471319 | 1.22 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr5_-_1325831 | 1.21 |
|
|
|
| chr16_-_32059848 | 1.18 |
ENSDART00000138701
|
gstk1
|
glutathione S-transferase kappa 1 |
| chr11_+_36983626 | 1.15 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr11_+_30482530 | 1.10 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr5_-_3562233 | 1.08 |
ENSDART00000143250
|
si:ch1073-189o9.1
|
si:ch1073-189o9.1 |
| chr13_-_22713071 | 1.07 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr7_-_64637051 | 1.07 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr15_-_23532475 | 1.05 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr13_-_37001997 | 1.05 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr13_+_45843396 | 1.05 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr11_+_11217547 | 1.05 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr14_+_24637864 | 1.04 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr8_-_11512545 | 1.03 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr24_+_20815964 | 1.02 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr21_-_12958161 | 1.01 |
ENSDART00000133517
|
MYORG
|
si:dkey-228b2.5 |
| chr16_-_12345194 | 1.00 |
ENSDART00000114759
|
lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr21_-_30074456 | 0.99 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr5_+_29327363 | 0.97 |
|
|
|
| chr2_+_37985514 | 0.97 |
ENSDART00000149224
|
apol1
|
apolipoprotein L, 1 |
| chr18_+_17622557 | 0.97 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr20_+_27194162 | 0.95 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr13_-_40600924 | 0.95 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr15_-_1880456 | 0.94 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr11_+_30049412 | 0.94 |
|
|
|
| chr5_+_29327479 | 0.93 |
|
|
|
| chr2_-_44429891 | 0.92 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr5_+_51392688 | 0.91 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr6_+_53428775 | 0.90 |
ENSDART00000165067
|
abhd14b
|
abhydrolase domain containing 14B |
| chr2_-_56531858 | 0.89 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr12_+_2412047 | 0.88 |
ENSDART00000112032
|
ARHGAP22 (1 of many)
|
Rho GTPase activating protein 22 |
| chr18_-_16934961 | 0.87 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr20_+_22766887 | 0.86 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr13_+_16130123 | 0.86 |
ENSDART00000170855
|
mocos
|
molybdenum cofactor sulfurase |
| chr25_+_3884499 | 0.84 |
ENSDART00000104926
|
ENSDARG00000058108
|
ENSDARG00000058108 |
| chr7_-_20201358 | 0.83 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr9_-_41982635 | 0.82 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr13_+_45843567 | 0.82 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr16_-_54638972 | 0.80 |
|
|
|
| chr17_-_16079935 | 0.80 |
|
|
|
| chr10_+_21693418 | 0.79 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr7_-_51186389 | 0.77 |
ENSDART00000174328
|
arhgap6
|
Rho GTPase activating protein 6 |
| chr9_+_8919542 | 0.76 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr2_-_37967102 | 0.74 |
ENSDART00000163165
|
CABZ01011067.1
|
ENSDARG00000105027 |
| chr11_-_30117563 | 0.74 |
ENSDART00000078387
|
pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr12_+_29994880 | 0.74 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr8_+_6533379 | 0.72 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| KN149817v1_+_2207 | 0.72 |
|
|
|
| chr23_-_3816492 | 0.70 |
ENSDART00000136394
|
hmga1a
|
high mobility group AT-hook 1a |
| chr18_-_7184884 | 0.69 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr14_-_32543646 | 0.69 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr20_+_22766918 | 0.68 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr7_-_20201693 | 0.66 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr7_+_30779987 | 0.66 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr8_+_39768744 | 0.66 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr22_+_24617667 | 0.65 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr6_-_2501966 | 0.65 |
ENSDART00000155109
|
hyi
|
hydroxypyruvate isomerase |
| chr5_-_44243476 | 0.65 |
ENSDART00000161408
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr10_+_33956052 | 0.64 |
|
|
|
| chr21_-_36886505 | 0.64 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr18_-_7441136 | 0.62 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr13_+_50062513 | 0.60 |
ENSDART00000124142
ENSDART00000099537 |
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr14_+_721560 | 0.60 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr11_+_6754335 | 0.60 |
ENSDART00000104292
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr21_-_37509396 | 0.59 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr24_-_36840203 | 0.59 |
ENSDART00000135142
|
si:ch73-334d15.1
|
si:ch73-334d15.1 |
| chr8_+_36888422 | 0.58 |
|
|
|
| chr4_-_19705018 | 0.58 |
ENSDART00000100974
|
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr8_+_13069277 | 0.56 |
ENSDART00000138433
|
itgb4
|
integrin, beta 4 |
| chr2_+_23600656 | 0.56 |
|
|
|
| chr23_-_5080531 | 0.56 |
|
|
|
| chr17_-_42778156 | 0.55 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr13_+_32676922 | 0.55 |
ENSDART00000085252
|
pqlc3
|
PQ loop repeat containing 3 |
| chr5_-_63487161 | 0.54 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr19_+_31944573 | 0.54 |
ENSDART00000136289
|
tmem64
|
transmembrane protein 64 |
| chr16_-_25691856 | 0.52 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr1_+_11485480 | 0.51 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr20_-_3074139 | 0.50 |
ENSDART00000046641
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr15_+_21399127 | 0.49 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr10_+_26786051 | 0.49 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr22_-_17570275 | 0.48 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr22_-_10744909 | 0.47 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr1_-_51658383 | 0.45 |
ENSDART00000132638
|
ENSDARG00000059948
|
ENSDARG00000059948 |
| chr20_-_3074186 | 0.44 |
ENSDART00000046641
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr10_-_17484971 | 0.44 |
|
|
|
| chr20_-_52526843 | 0.43 |
ENSDART00000132941
|
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr1_-_11377949 | 0.43 |
ENSDART00000103406
|
pla2g12a
|
phospholipase A2, group XIIA |
| chr10_+_10780578 | 0.42 |
ENSDART00000170147
ENSDART00000004181 |
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr12_-_944349 | 0.42 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr1_-_11377650 | 0.42 |
ENSDART00000143628
|
pla2g12a
|
phospholipase A2, group XIIA |
| chr12_+_16832804 | 0.42 |
|
|
|
| chr8_+_30690195 | 0.41 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr9_-_48152388 | 0.41 |
|
|
|
| chr9_-_470390 | 0.40 |
ENSDART00000162054
|
si:dkey-11f4.16
|
si:dkey-11f4.16 |
| chr18_-_47026460 | 0.40 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr1_+_14539766 | 0.39 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr20_+_5063644 | 0.39 |
ENSDART00000081351
ENSDART00000131103 ENSDART00000028039 |
cyp46a1.1
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 1 |
| chr18_+_18465885 | 0.36 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr7_-_73620490 | 0.36 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr8_+_47108500 | 0.36 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr3_+_24059652 | 0.36 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr2_+_3702282 | 0.35 |
ENSDART00000133007
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr23_-_45074750 | 0.35 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr15_-_29420984 | 0.35 |
ENSDART00000127795
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr5_+_29327447 | 0.34 |
|
|
|
| chr15_-_16140187 | 0.34 |
ENSDART00000080413
|
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr22_+_16293724 | 0.33 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr15_+_21399299 | 0.33 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr2_-_5816954 | 0.33 |
ENSDART00000014020
|
sst2
|
somatostatin 2 |
| chr3_-_49104540 | 0.31 |
ENSDART00000138503
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr1_-_45972732 | 0.30 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr13_-_30938325 | 0.30 |
ENSDART00000112653
|
wdfy4
|
WDFY family member 4 |
| chr23_-_24562107 | 0.30 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr24_-_32235674 | 0.29 |
ENSDART00000166212
|
cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr8_-_214434 | 0.29 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr4_-_16461881 | 0.29 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr6_-_40431708 | 0.29 |
ENSDART00000156005
|
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr12_+_29994657 | 0.28 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr15_-_1225639 | 0.27 |
|
|
|
| chr10_+_22004006 | 0.27 |
|
|
|
| chr24_+_36022833 | 0.26 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr5_+_43365449 | 0.25 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr23_+_17946933 | 0.25 |
ENSDART00000117743
|
SNORD59
|
Small nucleolar RNA SNORD59 |
| chr15_+_21808902 | 0.24 |
ENSDART00000016860
|
ppp2r1bb
|
protein phosphatase 2, regulatory subunit A, beta b |
| chr12_+_49091872 | 0.23 |
ENSDART00000178771
|
CABZ01075125.4
|
ENSDARG00000107503 |
| chr15_+_21399228 | 0.22 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr3_-_23513293 | 0.21 |
ENSDART00000140264
|
eve1
|
even-skipped-like1 |
| chr9_+_21572750 | 0.20 |
ENSDART00000130023
|
cryl1
|
crystallin, lambda 1 |
| chr17_-_17928974 | 0.19 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr23_-_9928561 | 0.19 |
|
|
|
| chr7_-_30009715 | 0.19 |
ENSDART00000173989
|
serf2
|
small EDRK-rich factor 2 |
| chr23_-_43703447 | 0.18 |
ENSDART00000170500
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr13_-_37002066 | 0.17 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr14_+_6122644 | 0.17 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr20_-_36906124 | 0.16 |
ENSDART00000166966
|
slc25a27
|
solute carrier family 25, member 27 |
| chr3_+_23536083 | 0.16 |
|
|
|
| chr8_+_39768702 | 0.14 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.6 | 1.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.5 | 1.5 | GO:1903793 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.5 | 3.9 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.5 | 2.8 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.4 | 0.4 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.4 | 3.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 3.2 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.4 | 3.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.4 | 1.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 2.0 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.3 | 1.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.3 | 1.6 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.2 | 1.0 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 1.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.2 | 1.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 0.7 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 4.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 2.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 0.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 0.6 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 1.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.7 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.2 | 2.9 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.2 | 0.7 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.2 | 1.7 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 6.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 2.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.7 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 2.9 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0098921 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.5 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 9.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 1.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.7 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.8 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 1.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.7 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 3.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.9 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.1 | GO:0072019 | proximal convoluted tubule development(GO:0072019) |
| 0.0 | 0.5 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 14.0 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.7 | 2.8 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.5 | 2.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 1.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 3.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 0.9 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 1.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 3.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.5 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.2 | 1.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 2.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 2.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 3.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 4.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.0 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 3.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 6.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 9.1 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 1.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 4.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 2.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 2.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 3.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |