Project
DANIO-CODE
Navigation
Downloads
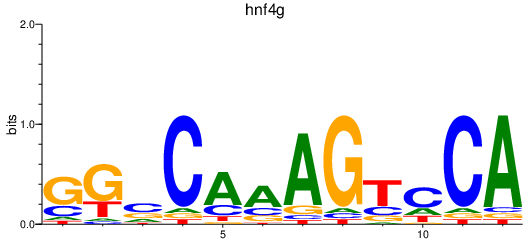
Results for hnf4g
Z-value: 1.29
Transcription factors associated with hnf4g
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf4g
|
ENSDARG00000071565 | hepatocyte nuclear factor 4, gamma |
Activity profile of hnf4g motif
Sorted Z-values of hnf4g motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf4g
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2401636 | 5.63 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2396632 | 4.89 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_+_26973049 | 4.62 |
ENSDART00000078124
|
trim35-29
|
tripartite motif containing 35-29 |
| chr22_-_24964889 | 4.13 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr14_+_44703094 | 3.91 |
|
|
|
| chr9_+_8402397 | 3.79 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr7_-_20358730 | 3.75 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr7_-_24249672 | 3.66 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr12_+_30389706 | 3.19 |
|
|
|
| chr19_-_33264349 | 2.86 |
|
|
|
| chr8_-_3354396 | 2.69 |
ENSDART00000169430
ENSDART00000167187 ENSDART00000170478 |
FUT9 (1 of many)
fut9b
|
fucosyltransferase 9 fucosyltransferase 9b |
| chr17_+_50622734 | 2.63 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr2_+_23004159 | 2.58 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr13_-_38666198 | 2.33 |
ENSDART00000162858
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr5_-_68408107 | 2.31 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr7_-_20358696 | 2.24 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr1_-_8801877 | 2.21 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr25_+_33118422 | 2.14 |
|
|
|
| chr15_+_29795467 | 1.99 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr7_-_24249630 | 1.98 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr3_-_36298676 | 1.93 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr17_-_2419079 | 1.92 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr15_+_34211736 | 1.92 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr24_-_7928026 | 1.69 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr9_+_29806055 | 1.69 |
ENSDART00000143493
|
phf11
|
PHD finger protein 11 |
| chr21_+_6890129 | 1.62 |
|
|
|
| chr23_+_20505691 | 1.61 |
ENSDART00000140219
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr1_-_11377907 | 1.60 |
ENSDART00000143628
|
pla2g12a
|
phospholipase A2, group XIIA |
| chr3_-_3665840 | 1.57 |
ENSDART00000078886
ENSDART00000170539 |
ENSDARG00000008835
|
ENSDARG00000008835 |
| chr9_-_5502510 | 1.55 |
ENSDART00000082260
|
abhd13
|
abhydrolase domain containing 13 |
| chr13_-_35782121 | 1.53 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr9_+_41658126 | 1.50 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr7_+_28453707 | 1.49 |
ENSDART00000173947
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr3_-_39920399 | 1.42 |
|
|
|
| chr5_-_35997345 | 1.33 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr5_-_56293700 | 1.32 |
|
|
|
| chr10_-_25737220 | 1.30 |
ENSDART00000135058
|
sod1
|
superoxide dismutase 1, soluble |
| chr15_+_29795509 | 1.29 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr7_-_20358897 | 1.27 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr5_+_32215942 | 1.26 |
ENSDART00000047377
|
crata
|
carnitine O-acetyltransferase a |
| chr17_-_2401568 | 1.23 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr14_-_16504730 | 1.21 |
ENSDART00000158396
|
tcirg1b
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3b |
| chr12_-_18839743 | 1.20 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr3_+_38497816 | 1.15 |
|
|
|
| chr11_-_27580252 | 1.15 |
ENSDART00000121847
|
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr7_+_28844711 | 1.15 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr21_-_10962154 | 1.13 |
ENSDART00000084061
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr2_-_48401770 | 1.13 |
ENSDART00000056277
|
gigfy2
|
GRB10 interacting GYF protein 2 |
| chr7_-_20358841 | 1.12 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr20_+_34248925 | 1.11 |
ENSDART00000145852
|
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr1_-_8801818 | 1.04 |
ENSDART00000008682
ENSDART00000157814 |
micall2b
|
mical-like 2b |
| chr5_+_26804676 | 1.04 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr7_-_26246669 | 1.00 |
ENSDART00000140694
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr13_+_26573404 | 0.95 |
ENSDART00000142483
|
fancl
|
Fanconi anemia, complementation group L |
| chr20_+_34248780 | 0.91 |
ENSDART00000152870
|
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr15_-_30976483 | 0.91 |
ENSDART00000112511
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr7_-_39467720 | 0.90 |
ENSDART00000052201
|
ccdc96
|
coiled-coil domain containing 96 |
| chr5_-_56294194 | 0.88 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr21_-_43478803 | 0.84 |
ENSDART00000151318
|
tmem185
|
transmembrane protein 185 |
| chr11_+_21949272 | 0.83 |
ENSDART00000122136
|
MDFI
|
MyoD family inhibitor |
| chr5_-_61654526 | 0.82 |
ENSDART00000158570
|
zgc:85789
|
zgc:85789 |
| chr12_-_18839808 | 0.76 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr22_+_132285 | 0.75 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr19_+_1356915 | 0.71 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr15_+_29795645 | 0.71 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr25_+_22119153 | 0.67 |
ENSDART00000051291
|
hexa
|
hexosaminidase A (alpha polypeptide) |
| chr15_-_25218118 | 0.65 |
ENSDART00000122184
|
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr9_+_41658313 | 0.61 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr10_+_22921629 | 0.61 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr2_-_11094083 | 0.60 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr15_-_16076807 | 0.60 |
ENSDART00000160114
|
hnf1ba
|
HNF1 homeobox Ba |
| chr17_-_49197264 | 0.59 |
ENSDART00000169207
|
tfb1m
|
transcription factor B1, mitochondrial |
| chr24_+_7798743 | 0.58 |
ENSDART00000134975
|
zgc:101569
|
zgc:101569 |
| chr5_+_23559189 | 0.58 |
ENSDART00000142268
|
gps2
|
G protein pathway suppressor 2 |
| chr7_-_56197660 | 0.57 |
|
|
|
| chr7_-_26246798 | 0.55 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr3_-_36298866 | 0.54 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr17_-_5918853 | 0.53 |
ENSDART00000058890
ENSDART00000171084 |
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| KN150363v1_+_14104 | 0.52 |
|
|
|
| chr5_-_23763146 | 0.49 |
|
|
|
| chr13_-_26573250 | 0.48 |
ENSDART00000139322
|
BX005477.1
|
ENSDARG00000093327 |
| chr19_-_1356587 | 0.44 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr6_-_54049582 | 0.44 |
|
|
|
| chr11_-_17882817 | 0.42 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr15_-_25218027 | 0.42 |
ENSDART00000078095
|
vps53
|
vacuolar protein sorting 53 homolog (S. cerevisiae) |
| chr24_-_7928168 | 0.39 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr9_+_38354060 | 0.38 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr18_+_11747188 | 0.38 |
ENSDART00000051105
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr11_-_27580123 | 0.34 |
ENSDART00000121847
|
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr15_+_8216629 | 0.33 |
ENSDART00000077660
|
mipep
|
mitochondrial intermediate peptidase |
| chr13_-_26573327 | 0.31 |
ENSDART00000141232
|
BX005477.1
|
ENSDARG00000093327 |
| chr7_+_39467483 | 0.30 |
ENSDART00000004365
|
tada2b
|
transcriptional adaptor 2B |
| chr22_+_1283662 | 0.29 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr3_+_45345291 | 0.29 |
|
|
|
| chr7_+_16977286 | 0.28 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr16_+_48518750 | 0.24 |
ENSDART00000162803
ENSDART00000159902 |
ext1a
|
exostosin glycosyltransferase 1a |
| chr5_+_43365528 | 0.19 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr11_+_24919823 | 0.18 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr1_+_14539897 | 0.17 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr7_-_28844560 | 0.12 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr8_+_39768702 | 0.11 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr15_+_8216822 | 0.09 |
ENSDART00000156442
|
mipep
|
mitochondrial intermediate peptidase |
| chr24_+_10942742 | 0.08 |
ENSDART00000136614
|
zfand1
|
zinc finger, AN1-type domain 1 |
| chr5_-_23650217 | 0.07 |
ENSDART00000080609
|
mrps31
|
mitochondrial ribosomal protein S31 |
| chr19_+_40535249 | 0.07 |
ENSDART00000151276
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr24_-_21740552 | 0.06 |
|
|
|
| chr11_+_42473839 | 0.06 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr6_-_33940469 | 0.04 |
ENSDART00000137268
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr21_-_11763422 | 0.04 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr11_+_30482504 | 0.04 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr15_-_16396862 | 0.01 |
ENSDART00000080892
|
txndc17
|
thioredoxin domain containing 17 |
| chr5_-_68408171 | 0.01 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.8 | 13.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 1.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.3 | 1.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.3 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 3.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 3.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 2.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 0.4 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 2.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.6 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 5.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.5 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 1.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 1.0 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 2.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 2.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 13.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 8.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.7 | GO:0005930 | axoneme(GO:0005930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.0 | 4.0 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.4 | 1.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.3 | 2.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 1.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.5 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 5.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 7.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 5.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |