Project
DANIO-CODE
Navigation
Downloads
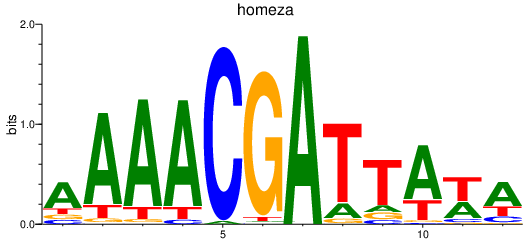
Results for homeza
Z-value: 1.32
Transcription factors associated with homeza
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
homeza
|
ENSDARG00000054304 | homeobox and leucine zipper encoding a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| homeza | dr10_dc_chr2_+_38288916_38289165 | 0.61 | 1.3e-02 | Click! |
Activity profile of homeza motif
Sorted Z-values of homeza motif
Network of associatons between targets according to the STRING database.
First level regulatory network of homeza
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34971985 | 3.49 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr6_-_8125341 | 3.18 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr21_-_13593659 | 3.01 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr21_+_20360180 | 2.78 |
ENSDART00000003299
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr15_-_20904022 | 2.59 |
ENSDART00000141746
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr1_-_51576354 | 2.48 |
ENSDART00000083946
|
pld6
|
phospholipase D family, member 6 |
| chr19_-_868378 | 2.38 |
|
|
|
| chr6_-_43297126 | 2.36 |
ENSDART00000154170
|
frmd4ba
|
FERM domain containing 4Ba |
| chr9_-_51745361 | 2.22 |
ENSDART00000154959
|
AL954669.1
|
ENSDARG00000097096 |
| chr7_+_17848688 | 2.18 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr16_+_27680365 | 2.16 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr10_-_25246786 | 2.08 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr8_-_23759076 | 2.01 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr7_+_15479700 | 2.01 |
|
|
|
| chr22_-_5600910 | 1.97 |
ENSDART00000179579
|
CABZ01075081.1
|
ENSDARG00000106226 |
| chr3_-_20833004 | 1.94 |
ENSDART00000160794
|
slc35b1
|
solute carrier family 35, member B1 |
| chr25_+_5845303 | 1.92 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr16_+_53391438 | 1.86 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr20_-_23527234 | 1.83 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_+_34183462 | 1.82 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr5_+_40896478 | 1.79 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr2_-_601945 | 1.78 |
|
|
|
| chr11_-_25019899 | 1.77 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr19_+_39689450 | 1.74 |
|
|
|
| chr2_+_27730940 | 1.74 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr2_-_57339717 | 1.71 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr5_+_69013399 | 1.67 |
ENSDART00000162057
ENSDART00000166893 |
setd8b
|
SET domain containing (lysine methyltransferase) 8b |
| chr9_-_46614763 | 1.65 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr10_+_14984467 | 1.64 |
|
|
|
| chr16_-_33047066 | 1.61 |
ENSDART00000147941
ENSDART00000075218 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr10_-_34971926 | 1.58 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr15_+_44168257 | 1.57 |
ENSDART00000176254
|
CU655961.7
|
ENSDARG00000106724 |
| chr2_+_11423028 | 1.57 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr6_-_12787256 | 1.54 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr11_-_40383013 | 1.54 |
ENSDART00000112140
|
fam213b
|
family with sequence similarity 213, member B |
| chr10_-_44634139 | 1.49 |
ENSDART00000010056
ENSDART00000169919 |
ccdc62
|
coiled-coil domain containing 62 |
| chr6_-_12786937 | 1.48 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr15_-_20903444 | 1.47 |
ENSDART00000139551
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr24_+_34183557 | 1.46 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| KN150226v1_+_11535 | 1.46 |
|
|
|
| chr3_-_20832951 | 1.40 |
ENSDART00000160794
|
slc35b1
|
solute carrier family 35, member B1 |
| chr19_+_8693855 | 1.38 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr15_-_970988 | 1.36 |
ENSDART00000106627
ENSDART00000102239 |
zgc:162936
|
zgc:162936 |
| chr3_+_16692138 | 1.35 |
ENSDART00000023985
|
zgc:153952
|
zgc:153952 |
| chr10_+_16078433 | 1.35 |
ENSDART00000065037
ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr3_-_32231069 | 1.34 |
ENSDART00000035545
|
prmt1
|
protein arginine methyltransferase 1 |
| chr10_-_44634167 | 1.34 |
ENSDART00000010056
ENSDART00000169919 |
ccdc62
|
coiled-coil domain containing 62 |
| chr8_+_41004169 | 1.33 |
|
|
|
| chr8_+_40463218 | 1.33 |
|
|
|
| chr24_+_39237314 | 1.32 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr15_-_20904050 | 1.32 |
ENSDART00000141746
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr3_+_30187036 | 1.31 |
ENSDART00000151006
|
CR936968.1
|
ENSDARG00000096295 |
| chr10_+_16078021 | 1.31 |
ENSDART00000065037
ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr24_+_23571714 | 1.28 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr15_+_24741620 | 1.27 |
ENSDART00000078014
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr5_+_40896320 | 1.27 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr21_+_20360748 | 1.27 |
ENSDART00000146615
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr8_-_22493608 | 1.26 |
ENSDART00000021514
|
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr8_-_11806733 | 1.26 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr18_-_35432838 | 1.25 |
ENSDART00000141703
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr8_-_29842489 | 1.24 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr6_-_8125287 | 1.24 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr6_-_6818607 | 1.24 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr3_-_19913207 | 1.23 |
|
|
|
| chr15_+_24628191 | 1.22 |
ENSDART00000140658
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr17_+_28594742 | 1.22 |
ENSDART00000062430
ENSDART00000113300 |
mis18bp1
|
MIS18 binding protein 1 |
| chr5_-_31257158 | 1.22 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr25_+_20596490 | 1.20 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr3_-_39117855 | 1.20 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| chr18_-_43890836 | 1.19 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr17_+_16184621 | 1.17 |
ENSDART00000156832
|
kif13ba
|
kinesin family member 13Ba |
| chr4_+_13902603 | 1.17 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr16_-_12582192 | 1.17 |
|
|
|
| chr17_-_24860924 | 1.15 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr13_+_3807780 | 1.14 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr8_-_11806698 | 1.14 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr16_+_53632289 | 1.13 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr1_-_29958364 | 1.13 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr18_-_35432655 | 1.13 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr23_+_27777325 | 1.11 |
ENSDART00000134008
|
lmbr1l
|
limb development membrane protein 1-like |
| chr4_+_9010972 | 1.11 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr5_+_29996321 | 1.11 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr14_-_30196413 | 1.09 |
ENSDART00000178574
|
FP015860.1
|
ENSDARG00000106816 |
| chr19_-_31988686 | 1.09 |
ENSDART00000161739
|
tdp2b
|
tyrosyl-DNA phosphodiesterase 2b |
| chr24_-_21224371 | 1.08 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr2_-_19705537 | 1.07 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr23_+_35327654 | 1.07 |
ENSDART00000165034
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr3_+_18287783 | 1.07 |
ENSDART00000144690
|
tbc1d16
|
TBC1 domain family, member 16 |
| chr7_+_39812522 | 1.06 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr18_+_22426900 | 1.05 |
ENSDART00000115388
|
cfdp1
|
craniofacial development protein 1 |
| chr7_+_22524917 | 1.04 |
ENSDART00000112169
|
rbm4.2
|
RNA binding motif protein 4.2 |
| chr15_-_20903489 | 1.03 |
ENSDART00000139551
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| KN150226v1_+_11474 | 1.03 |
|
|
|
| chr24_+_35296732 | 1.03 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr15_+_46053808 | 1.03 |
|
|
|
| chr1_-_25970424 | 1.02 |
ENSDART00000152401
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr21_-_43654780 | 1.01 |
ENSDART00000142733
|
si:dkey-229d11.5
|
si:dkey-229d11.5 |
| chr18_-_12358605 | 1.01 |
ENSDART00000114024
|
fam107b
|
family with sequence similarity 107, member B |
| chr1_-_25369230 | 1.00 |
ENSDART00000171752
|
pdcd4a
|
programmed cell death 4a |
| chr23_+_6652454 | 0.99 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr13_-_44885278 | 0.99 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr7_+_39812659 | 0.99 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr1_-_51075777 | 0.99 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr7_-_64745132 | 0.98 |
ENSDART00000112166
|
fam60al
|
family with sequence similarity 60, member A, like |
| chr24_+_28304276 | 0.98 |
ENSDART00000018095
|
sh3glb1a
|
SH3-domain GRB2-like endophilin B1a |
| chr8_-_39788989 | 0.98 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr20_-_49067167 | 0.98 |
ENSDART00000163071
ENSDART00000170617 |
xrn2
|
5'-3' exoribonuclease 2 |
| chr23_+_33981443 | 0.97 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr11_+_19440815 | 0.97 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr19_+_7082199 | 0.97 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr12_+_6197452 | 0.97 |
|
|
|
| chr12_-_4800362 | 0.96 |
ENSDART00000172093
|
si:ch211-93e11.8
|
si:ch211-93e11.8 |
| chr15_+_17350036 | 0.96 |
ENSDART00000134512
|
cltcb
|
clathrin, heavy chain b (Hc) |
| chr21_+_11686037 | 0.95 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr20_+_1364879 | 0.95 |
ENSDART00000145981
ENSDART00000152709 |
mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr8_-_16689736 | 0.95 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr11_+_14285767 | 0.95 |
ENSDART00000171347
|
si:ch211-262i1.6
|
si:ch211-262i1.6 |
| chr3_+_18287645 | 0.94 |
ENSDART00000136243
|
tbc1d16
|
TBC1 domain family, member 16 |
| chr4_+_29008199 | 0.94 |
ENSDART00000150629
|
si:dkey-23a23.3
|
si:dkey-23a23.3 |
| chr24_-_21224397 | 0.94 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr9_+_8990774 | 0.94 |
ENSDART00000133899
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr1_+_40894908 | 0.91 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr7_+_66410533 | 0.91 |
ENSDART00000027616
ENSDART00000162763 |
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr14_-_23980060 | 0.91 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr15_+_24741931 | 0.91 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr14_+_14850200 | 0.91 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr3_+_30962827 | 0.91 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr18_-_6896957 | 0.91 |
ENSDART00000175747
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr16_+_29090602 | 0.91 |
ENSDART00000149454
|
gpatch4
|
G patch domain containing 4 |
| chr16_-_29470341 | 0.90 |
ENSDART00000149289
|
tlr18
|
toll-like receptor 18 |
| chr19_+_42758938 | 0.90 |
ENSDART00000077059
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr25_+_20596613 | 0.90 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr15_-_20903583 | 0.90 |
ENSDART00000031625
|
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr22_+_17474904 | 0.90 |
ENSDART00000061366
|
rab11ba
|
RAB11B, member RAS oncogene family, a |
| chr20_+_40590886 | 0.89 |
|
|
|
| chr9_-_41238226 | 0.89 |
ENSDART00000008275
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr20_+_6545449 | 0.88 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr23_-_31323662 | 0.88 |
|
|
|
| chr25_-_15176295 | 0.87 |
|
|
|
| KN150712v1_-_26150 | 0.87 |
|
|
|
| chr24_+_23571827 | 0.87 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| KN150034v1_+_1223 | 0.86 |
|
|
|
| chr7_+_39812732 | 0.86 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr9_+_44921195 | 0.86 |
ENSDART00000145271
|
nckap1
|
NCK-associated protein 1 |
| chr19_-_17400440 | 0.86 |
ENSDART00000160433
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr9_-_20743808 | 0.86 |
|
|
|
| chr13_-_36486255 | 0.85 |
ENSDART00000160250
|
BX927407.1
|
ENSDARG00000103360 |
| chr8_-_16479425 | 0.85 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr10_-_36849596 | 0.85 |
|
|
|
| chr8_-_20882626 | 0.85 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr21_-_11106675 | 0.84 |
ENSDART00000167666
|
dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr17_+_23534953 | 0.84 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr13_+_3808095 | 0.84 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr17_-_15487267 | 0.84 |
|
|
|
| chr3_+_30962542 | 0.83 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr19_+_7708513 | 0.83 |
ENSDART00000129994
ENSDART00000131324 ENSDART00000104731 |
pygo2
|
pygopus homolog 2 (Drosophila) |
| chr10_-_15382145 | 0.82 |
ENSDART00000133919
|
pum3
|
pumilio RNA-binding family member 3 |
| chr7_-_18460264 | 0.82 |
ENSDART00000173905
|
trmt10a
|
tRNA methyltransferase 10A |
| chr19_-_4206143 | 0.82 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr9_+_8990576 | 0.82 |
ENSDART00000133899
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr5_+_37497397 | 0.82 |
|
|
|
| chr9_-_10174409 | 0.81 |
ENSDART00000004745
|
hnmt
|
histamine N-methyltransferase |
| chr9_-_25370812 | 0.81 |
ENSDART00000012582
|
esd
|
esterase D/formylglutathione hydrolase |
| chr6_-_52677363 | 0.81 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr17_+_13292569 | 0.79 |
ENSDART00000177104
|
BX005308.2
|
ENSDARG00000108498 |
| chr16_-_31494275 | 0.79 |
ENSDART00000056551
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr25_-_31979408 | 0.78 |
|
|
|
| chr1_-_25970563 | 0.78 |
ENSDART00000152401
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr10_-_36849734 | 0.78 |
|
|
|
| chr22_-_6916274 | 0.78 |
|
|
|
| chr9_-_34700736 | 0.77 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr3_+_40147509 | 0.77 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr23_-_41367783 | 0.77 |
|
|
|
| chr22_-_24957034 | 0.77 |
ENSDART00000142147
|
dnal4b
|
dynein, axonemal, light chain 4b |
| chr3_+_52699683 | 0.77 |
ENSDART00000125136
|
dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr3_+_19490148 | 0.76 |
|
|
|
| chr21_-_21604542 | 0.75 |
ENSDART00000010282
|
cbr1l
|
carbonyl reductase 1-like |
| chr25_-_12316228 | 0.75 |
ENSDART00000168275
|
det1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr22_-_9809500 | 0.75 |
|
|
|
| chr3_+_22914688 | 0.74 |
ENSDART00000156472
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr7_+_28341426 | 0.74 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr11_+_37385705 | 0.74 |
ENSDART00000129918
|
kif17
|
kinesin family member 17 |
| chr9_+_55429357 | 0.74 |
|
|
|
| chr20_+_44600015 | 0.74 |
ENSDART00000023763
|
wdcp
|
WD repeat and coiled coil containing |
| chr19_+_8693695 | 0.74 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr19_-_27755498 | 0.74 |
ENSDART00000137346
|
znrd1
|
zinc ribbon domain containing 1 |
| chr15_+_20303673 | 0.73 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr8_-_44247277 | 0.73 |
|
|
|
| chr12_-_308764 | 0.73 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr8_-_8451037 | 0.73 |
ENSDART00000064113
|
abt1
|
activator of basal transcription 1 |
| chr16_+_53632152 | 0.71 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr3_+_35292809 | 0.71 |
ENSDART00000176673
|
BX255942.1
|
ENSDARG00000108159 |
| chr9_+_32838316 | 0.71 |
ENSDART00000172033
|
rabl3
|
RAB, member of RAS oncogene family-like 3 |
| chr6_-_24292584 | 0.71 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr12_-_31346300 | 0.71 |
ENSDART00000175929
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr14_-_32319519 | 0.71 |
|
|
|
| chr7_-_51474798 | 0.71 |
ENSDART00000175523
|
hdac8
|
histone deacetylase 8 |
| chr3_-_26112995 | 0.71 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr1_-_25369054 | 0.70 |
ENSDART00000171752
|
pdcd4a
|
programmed cell death 4a |
| chr16_+_54953101 | 0.70 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.6 | 4.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.5 | 1.9 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.5 | 1.4 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.4 | 3.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 1.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 2.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 1.4 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.3 | 3.0 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 | 2.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.3 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.3 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 1.2 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.3 | 1.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 0.8 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 | 1.9 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 0.8 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 2.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.2 | 0.7 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) RNA 5'-end processing(GO:0000966) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 1.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.7 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.7 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 0.6 | GO:0071887 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.2 | 1.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.7 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.2 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 0.5 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.2 | 2.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.2 | 0.8 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 1.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 2.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.0 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 1.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 7.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 0.7 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.8 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 1.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.4 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.3 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 2.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.1 | 0.2 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.1 | 0.5 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.7 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 5.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.8 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.6 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.3 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.0 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 1.7 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.0 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.3 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 1.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.6 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.1 | 0.3 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.2 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.0 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.7 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.1 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0034727 | lysosomal microautophagy(GO:0016237) pexophagy(GO:0030242) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.8 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.4 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.3 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.1 | GO:0065001 | specification of organ axis polarity(GO:0010084) specification of axis polarity(GO:0065001) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.5 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.6 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 6.5 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.0 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.1 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.2 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.6 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 3.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 2.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 2.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 0.8 | GO:0000941 | nuclear MIS12/MIND complex(GO:0000818) condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.3 | 1.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 2.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 3.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 2.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.7 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.8 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.9 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 2.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 5.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 3.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0031968 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.5 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 1.4 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0035755 | cardiolipin hydrolase activity(GO:0035755) |
| 0.8 | 7.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.6 | 2.5 | GO:0005460 | UDP-glucose transmembrane transporter activity(GO:0005460) |
| 0.6 | 4.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.5 | 1.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.5 | 1.8 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.4 | 1.3 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.4 | 1.7 | GO:0070260 | 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.4 | 1.3 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.4 | 3.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 3.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 1.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 1.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.2 | 1.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.6 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 1.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.7 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 1.9 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.2 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 1.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 1.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.4 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.6 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.6 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.4 | GO:0030882 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.1 | 0.3 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 1.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 5.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.3 | GO:0061133 | peptidase activator activity(GO:0016504) endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 1.0 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 6.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 4.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |