Project
DANIO-CODE
Navigation
Downloads
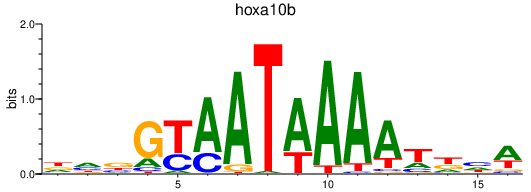
Results for hoxa10b
Z-value: 0.82
Transcription factors associated with hoxa10b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa10b
|
ENSDARG00000031337 | homeobox A10b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa10b | dr10_dc_chr16_+_21104310_21104375 | -0.97 | 7.5e-10 | Click! |
Activity profile of hoxa10b motif
Sorted Z-values of hoxa10b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa10b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_25019899 | 4.21 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr7_+_1337856 | 2.21 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr10_+_15382647 | 1.85 |
ENSDART00000046274
|
trappc13
|
trafficking protein particle complex 13 |
| chr2_-_21512154 | 1.83 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr11_-_25019298 | 1.83 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr12_-_10474942 | 1.79 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr15_-_43954665 | 1.79 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr10_-_21587697 | 1.78 |
ENSDART00000029122
|
zgc:165539
|
zgc:165539 |
| chr1_-_52833379 | 1.76 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr25_+_8936284 | 1.75 |
ENSDART00000154207
|
im:7145024
|
im:7145024 |
| chr4_+_4825461 | 1.75 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr19_-_4206333 | 1.74 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr21_+_43183522 | 1.70 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr5_-_31257158 | 1.62 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr23_+_28563324 | 1.61 |
ENSDART00000006657
|
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr14_-_7102535 | 1.60 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr14_-_880799 | 1.59 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr22_+_1837102 | 1.58 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr5_-_57053687 | 1.55 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr17_+_50622734 | 1.55 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| KN150334v1_-_9585 | 1.51 |
ENSDART00000175935
|
CABZ01113810.1
|
ENSDARG00000107898 |
| chr2_-_30929469 | 1.43 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr5_-_23211957 | 1.40 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr3_-_29779725 | 1.38 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr8_-_37989930 | 1.30 |
|
|
|
| chr24_-_23639458 | 1.29 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr3_-_29779598 | 1.25 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr15_+_14656797 | 1.20 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr8_+_23083842 | 1.19 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr2_+_35750334 | 1.17 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr4_+_16020464 | 1.16 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr23_-_35691369 | 1.12 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr17_-_30635487 | 1.05 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr7_+_40878834 | 1.05 |
|
|
|
| chr13_-_24586813 | 1.03 |
ENSDART00000137934
|
erlin1
|
ER lipid raft associated 1 |
| chr21_-_2273151 | 1.02 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr14_-_15777250 | 1.02 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr8_+_36527653 | 1.01 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr17_+_15527525 | 0.98 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr10_-_25448712 | 0.98 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr23_-_27579257 | 0.95 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr3_+_6557967 | 0.93 |
ENSDART00000162255
|
nup85
|
nucleoporin 85 |
| chr14_+_10319183 | 0.93 |
ENSDART00000136480
|
nup62l
|
nucleoporin 62 like |
| chr23_-_29825098 | 0.92 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr24_-_23639592 | 0.91 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr21_-_2273243 | 0.91 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr18_-_7138350 | 0.89 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr8_-_18869085 | 0.88 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr25_-_29020637 | 0.87 |
ENSDART00000124645
|
ube2q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr5_-_64432697 | 0.86 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr3_+_12087549 | 0.84 |
|
|
|
| chr11_+_11319810 | 0.84 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr3_-_29779552 | 0.83 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr3_+_13450981 | 0.83 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr23_-_24328931 | 0.82 |
|
|
|
| chr13_+_13814142 | 0.82 |
ENSDART00000142997
|
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr17_+_17745119 | 0.81 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr19_-_6925090 | 0.81 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr9_+_34423285 | 0.81 |
ENSDART00000174944
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr6_-_3763940 | 0.80 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr4_+_4825409 | 0.80 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr6_-_9046536 | 0.80 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr18_+_1778654 | 0.79 |
ENSDART00000176491
|
CABZ01073163.1
|
ENSDARG00000107400 |
| chr23_+_20505072 | 0.79 |
ENSDART00000132920
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr21_-_26655258 | 0.79 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_8801818 | 0.79 |
ENSDART00000008682
ENSDART00000157814 |
micall2b
|
mical-like 2b |
| chr13_+_1001437 | 0.75 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr12_-_18839743 | 0.75 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr13_+_8560739 | 0.74 |
ENSDART00000075054
|
thada
|
thyroid adenoma associated |
| chr20_+_25687135 | 0.74 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr6_+_4091120 | 0.73 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr3_+_39437497 | 0.73 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr1_-_13302923 | 0.73 |
ENSDART00000011414
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr3_+_39437613 | 0.73 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr1_-_52833426 | 0.73 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr1_-_11707244 | 0.73 |
ENSDART00000146067
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr2_+_22753634 | 0.72 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr12_-_16403282 | 0.72 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr11_-_5956254 | 0.72 |
ENSDART00000140960
|
dda1
|
DET1 and DDB1 associated 1 |
| chr10_-_31861975 | 0.72 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr15_+_14656886 | 0.71 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr14_-_44880144 | 0.71 |
ENSDART00000163543
|
abhd18
|
abhydrolase domain containing 18 |
| chr17_+_15527861 | 0.70 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr12_+_13167781 | 0.70 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr1_+_11707382 | 0.68 |
ENSDART00000139440
|
zgc:77739
|
zgc:77739 |
| chr5_-_31257031 | 0.67 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr13_-_36486255 | 0.67 |
ENSDART00000160250
|
BX927407.1
|
ENSDARG00000103360 |
| chr18_-_7138309 | 0.67 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr2_-_30929321 | 0.65 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr2_-_41035670 | 0.65 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr13_-_17729510 | 0.64 |
ENSDART00000170583
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr25_-_28630823 | 0.63 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr17_-_30635724 | 0.63 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr12_+_13167838 | 0.62 |
ENSDART00000160664
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr10_-_20631301 | 0.62 |
ENSDART00000138045
|
whsc1l1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr15_-_23973374 | 0.61 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr18_-_7138171 | 0.61 |
ENSDART00000149122
|
cfap161
|
cilia and flagella associated protein 161 |
| chr13_+_7243353 | 0.60 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr14_-_31513 | 0.60 |
ENSDART00000161122
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr22_-_20925422 | 0.60 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr11_-_1382526 | 0.60 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr20_-_34865344 | 0.59 |
ENSDART00000142482
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr23_-_32464136 | 0.58 |
ENSDART00000032861
|
ankrd52a
|
ankyrin repeat domain 52a |
| chr19_-_22762165 | 0.58 |
ENSDART00000169065
ENSDART00000080260 |
zgc:109744
|
zgc:109744 |
| chr18_+_20058174 | 0.58 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr13_-_24586850 | 0.57 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr10_-_20631240 | 0.56 |
ENSDART00000138045
|
whsc1l1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr10_+_6011791 | 0.56 |
ENSDART00000159216
|
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr21_-_18909848 | 0.56 |
ENSDART00000139622
|
med15
|
mediator complex subunit 15 |
| chr11_-_25019165 | 0.55 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr2_-_41035732 | 0.55 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr20_-_45806178 | 0.55 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr3_+_47826595 | 0.55 |
ENSDART00000155409
|
unkl
|
unkempt family zinc finger-like |
| chr6_-_6818607 | 0.54 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr18_-_46024614 | 0.54 |
|
|
|
| chr20_+_17682270 | 0.54 |
|
|
|
| chr13_-_27223507 | 0.53 |
ENSDART00000101479
|
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr14_+_37506462 | 0.52 |
ENSDART00000174790
ENSDART00000179610 ENSDART00000179048 |
CU104743.1
|
ENSDARG00000109062 |
| KN149795v1_-_10712 | 0.52 |
|
|
|
| chr8_-_51537679 | 0.51 |
ENSDART00000147742
|
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr18_-_50617567 | 0.51 |
ENSDART00000172530
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr12_+_11314303 | 0.51 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr9_+_2336680 | 0.51 |
ENSDART00000016417
|
atp5g3a
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9), genome duplicate a |
| chr4_-_27166180 | 0.50 |
|
|
|
| chr4_-_11054645 | 0.50 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr7_+_36444234 | 0.50 |
|
|
|
| chr20_-_7079412 | 0.50 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr2_+_22753590 | 0.49 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr8_+_2597651 | 0.49 |
ENSDART00000165943
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr14_+_22170431 | 0.49 |
ENSDART00000079409
|
nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr24_-_33915389 | 0.48 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr13_+_13814262 | 0.48 |
ENSDART00000142997
|
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr23_-_1562603 | 0.47 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr6_-_19794771 | 0.46 |
ENSDART00000151444
|
setd5
|
SET domain containing 5 |
| chr21_-_11877525 | 0.46 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr2_-_11094083 | 0.45 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr10_-_31862004 | 0.44 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr19_+_40482359 | 0.44 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr20_-_45806016 | 0.44 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr22_+_2313616 | 0.44 |
|
|
|
| chr4_+_4825628 | 0.43 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr10_-_10755962 | 0.43 |
ENSDART00000137797
|
CT027791.1
|
ENSDARG00000093881 |
| chr18_+_21962395 | 0.43 |
ENSDART00000089507
|
ranbp10
|
RAN binding protein 10 |
| chr13_+_7243325 | 0.43 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr1_-_55195566 | 0.42 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr19_+_2942092 | 0.42 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr19_-_31212648 | 0.41 |
ENSDART00000125893
ENSDART00000145581 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr14_+_21835107 | 0.41 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr5_-_67395562 | 0.39 |
ENSDART00000165052
ENSDART00000018792 |
spag7
|
sperm associated antigen 7 |
| chr15_-_1880173 | 0.38 |
ENSDART00000154175
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr16_+_31988496 | 0.38 |
ENSDART00000058496
|
phc1
|
polyhomeotic homolog 1 |
| chr18_-_7138414 | 0.38 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr9_+_2538387 | 0.37 |
|
|
|
| chr3_+_15209283 | 0.37 |
ENSDART00000143280
|
sh2b1
|
SH2B adaptor protein 1 |
| chr15_-_1768232 | 0.37 |
ENSDART00000023153
|
rabgef1l
|
RAB guanine nucleotide exchange factor (GEF) 1, like |
| chr21_-_13133214 | 0.37 |
|
|
|
| chr19_-_44469947 | 0.37 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr14_-_880729 | 0.36 |
ENSDART00000165211
|
rgs14a
|
regulator of G protein signaling 14a |
| chr17_-_30504063 | 0.36 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr15_-_1768269 | 0.36 |
ENSDART00000154668
|
rabgef1l
|
RAB guanine nucleotide exchange factor (GEF) 1, like |
| chr15_+_11772499 | 0.35 |
|
|
|
| chr6_+_40556891 | 0.35 |
ENSDART00000155928
|
ddi2
|
DNA-damage inducible protein 2 |
| chr16_-_6304889 | 0.35 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr11_-_5956172 | 0.34 |
ENSDART00000140960
|
dda1
|
DET1 and DDB1 associated 1 |
| chr4_-_11054766 | 0.34 |
ENSDART00000161470
|
mettl25
|
methyltransferase like 25 |
| chr2_+_30976732 | 0.33 |
ENSDART00000141575
|
lipin2
|
lipin 2 |
| chr21_+_812425 | 0.33 |
ENSDART00000006419
ENSDART00000133976 |
txnl1
|
thioredoxin-like 1 |
| chr18_+_45866556 | 0.32 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr12_-_18839808 | 0.31 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr9_-_54459836 | 0.30 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr23_+_36208651 | 0.30 |
ENSDART00000127174
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr23_-_29579049 | 0.30 |
ENSDART00000165744
|
kif1b
|
kinesin family member 1B |
| chr16_-_51434927 | 0.30 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr3_+_13450861 | 0.29 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr15_-_47341913 | 0.29 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr15_-_43954596 | 0.29 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr13_-_44883668 | 0.29 |
|
|
|
| chr15_-_45304762 | 0.29 |
ENSDART00000177018
|
CABZ01068243.1
|
ENSDARG00000106883 |
| chr8_-_6782167 | 0.29 |
ENSDART00000135834
ENSDART00000172157 |
dock5
|
dedicator of cytokinesis 5 |
| chr22_+_39067938 | 0.28 |
|
|
|
| chr5_+_23559435 | 0.28 |
ENSDART00000142268
|
gps2
|
G protein pathway suppressor 2 |
| chr3_+_39514432 | 0.28 |
ENSDART00000156038
|
epn2
|
epsin 2 |
| chr19_+_28386051 | 0.28 |
ENSDART00000103887
|
med10
|
mediator complex subunit 10 |
| chr16_+_38387709 | 0.28 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr3_+_40434532 | 0.27 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr1_+_56656523 | 0.27 |
ENSDART00000152229
|
ENSDARG00000007289
|
ENSDARG00000007289 |
| chr20_-_2624028 | 0.27 |
ENSDART00000145335
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr14_+_10319051 | 0.26 |
ENSDART00000136480
|
nup62l
|
nucleoporin 62 like |
| chr12_-_16403608 | 0.26 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr10_+_39142185 | 0.26 |
|
|
|
| chr15_+_17322173 | 0.26 |
ENSDART00000169608
|
dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr25_-_3168287 | 0.25 |
|
|
|
| chr15_-_23973415 | 0.25 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr15_+_23598869 | 0.25 |
ENSDART00000152320
|
si:dkey-182i3.10
|
si:dkey-182i3.10 |
| chr21_-_14781371 | 0.24 |
ENSDART00000131237
|
ulk1b
|
unc-51 like autophagy activating kinase 1 |
| chr18_-_50617626 | 0.23 |
ENSDART00000172530
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr14_+_34626191 | 0.23 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr3_+_15743134 | 0.23 |
ENSDART00000114564
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr21_+_43706830 | 0.22 |
ENSDART00000126190
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr20_-_51915872 | 0.22 |
ENSDART00000004092
|
brox
|
BRO1 domain and CAAX motif containing |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1900144 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.5 | 2.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 1.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 1.6 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 0.8 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 | 6.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.5 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) skin morphogenesis(GO:0043589) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 0.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 1.6 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 1.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 3.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.5 | GO:0061698 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 1.3 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 1.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 1.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.3 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 1.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.7 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 0.2 | GO:1990481 | snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.2 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.1 | 1.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 2.1 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 1.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 1.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 4.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.0 | 0.5 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 1.1 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 1.7 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.9 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 1.8 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.6 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 1.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 1.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 0.8 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 1.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.1 | 2.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 1.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 2.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.1 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.0 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 6.2 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.3 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 3.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |