Project
DANIO-CODE
Navigation
Downloads
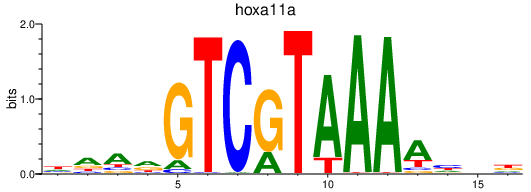
Results for hoxa11a
Z-value: 0.80
Transcription factors associated with hoxa11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11a
|
ENSDARG00000104162 | homeobox A11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11a | dr10_dc_chr19_+_20164849_20164864 | 0.37 | 1.6e-01 | Click! |
Activity profile of hoxa11a motif
Sorted Z-values of hoxa11a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_43998886 | 2.12 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr21_+_13031158 | 1.93 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr1_-_23765358 | 1.86 |
|
|
|
| chr7_-_18256512 | 1.85 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr2_-_3317258 | 1.60 |
ENSDART00000167944
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr12_-_26760324 | 1.47 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr21_+_13030816 | 1.44 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr9_-_3700395 | 1.25 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr11_-_2107054 | 1.09 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr7_+_20272091 | 1.08 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr15_-_14616083 | 0.95 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr23_+_35988395 | 0.95 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr2_-_10555152 | 0.90 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr25_+_17906515 | 0.87 |
|
|
|
| chr10_+_2559774 | 0.86 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr23_+_35988426 | 0.84 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr15_-_18110169 | 0.77 |
|
|
|
| chr3_+_36155364 | 0.69 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr21_+_13031206 | 0.65 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr12_+_24833569 | 0.65 |
ENSDART00000014868
|
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr5_+_36010448 | 0.65 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr25_-_17906739 | 0.65 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr25_-_17906512 | 0.62 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr1_+_30215209 | 0.61 |
ENSDART00000164520
|
itga6b
|
integrin, alpha 6b |
| chr7_-_18256590 | 0.58 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr17_-_23707863 | 0.54 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr14_+_30568242 | 0.53 |
|
|
|
| chr7_+_20699940 | 0.52 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr22_+_36562719 | 0.52 |
ENSDART00000178537
|
CABZ01045213.1
|
ENSDARG00000107720 |
| chr25_+_17906724 | 0.50 |
|
|
|
| chr16_-_10425382 | 0.49 |
ENSDART00000104025
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr23_+_35988366 | 0.47 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr12_+_33261056 | 0.47 |
ENSDART00000124982
|
ccdc57
|
coiled-coil domain containing 57 |
| chr5_+_58727722 | 0.46 |
ENSDART00000082983
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr6_+_7093685 | 0.45 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr12_+_27026112 | 0.44 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr25_-_20994084 | 0.41 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr5_+_58727940 | 0.40 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr6_+_50394326 | 0.39 |
ENSDART00000055511
ENSDART00000165063 |
ergic3
|
ERGIC and golgi 3 |
| chr16_-_6304533 | 0.39 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr17_-_31678247 | 0.36 |
ENSDART00000143090
|
lin52
|
lin-52 DREAM MuvB core complex component |
| chr20_-_46638031 | 0.35 |
ENSDART00000060685
|
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr7_-_40734662 | 0.32 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr9_+_20672966 | 0.30 |
ENSDART00000145111
|
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr2_-_37891963 | 0.27 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr15_+_8791540 | 0.25 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr7_+_20700070 | 0.23 |
|
|
|
| chr16_-_47491350 | 0.23 |
ENSDART00000149723
|
sept7b
|
septin 7b |
| chr12_-_34612758 | 0.21 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr19_+_7505348 | 0.20 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr13_-_9543393 | 0.16 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr19_-_18671902 | 0.16 |
|
|
|
| chr3_+_24059652 | 0.16 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr7_+_9063087 | 0.15 |
ENSDART00000084598
|
vimp
|
VCP-interacting membrane selenoprotein |
| chr11_-_35788900 | 0.14 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr25_-_20993989 | 0.12 |
|
|
|
| chr7_-_30009715 | 0.09 |
ENSDART00000173989
|
serf2
|
small EDRK-rich factor 2 |
| chr23_+_35980812 | 0.07 |
|
|
|
| chr22_-_33942334 | 0.06 |
|
|
|
| chr24_+_20514498 | 0.04 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr7_-_30353264 | 0.04 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr24_-_21740552 | 0.03 |
|
|
|
| chr1_-_9256864 | 0.00 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 0.5 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.2 | 1.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.1 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.4 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.5 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.2 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 3.3 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.4 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 4.5 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |