Project
DANIO-CODE
Navigation
Downloads
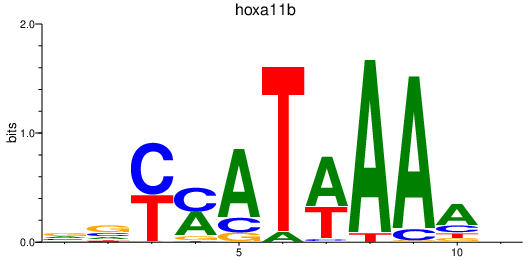
Results for hoxa11b
Z-value: 2.30
Transcription factors associated with hoxa11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11b
|
ENSDARG00000007009 | homeobox A11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11b | dr10_dc_chr16_+_21098948_21099031 | -0.89 | 3.8e-06 | Click! |
Activity profile of hoxa11b motif
Sorted Z-values of hoxa11b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_40718244 | 9.32 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr13_-_28480530 | 8.90 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr14_+_34150130 | 8.58 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr19_-_27966525 | 8.03 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr21_-_32747592 | 6.28 |
|
|
|
| chr12_+_22552867 | 5.69 |
ENSDART00000152930
|
cdca9
|
cell division cycle associated 9 |
| chr7_+_1337856 | 5.51 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr2_+_50680126 | 5.48 |
ENSDART00000122716
|
ENSDARG00000090398
|
ENSDARG00000090398 |
| chr21_+_20360180 | 5.43 |
ENSDART00000003299
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr6_+_28218420 | 5.34 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr5_-_23211957 | 5.18 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr5_-_53907908 | 5.11 |
ENSDART00000158069
|
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr14_-_880799 | 4.61 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr20_-_40357302 | 4.59 |
|
|
|
| chr1_+_41644371 | 4.53 |
ENSDART00000143871
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr12_+_33257120 | 4.46 |
|
|
|
| chr8_+_45326435 | 4.28 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr7_+_15479700 | 4.18 |
|
|
|
| chr22_+_1837102 | 4.02 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr3_-_26052785 | 3.95 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr15_+_29092022 | 3.95 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr21_+_43183522 | 3.93 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr16_+_29574449 | 3.89 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr1_-_52833379 | 3.89 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr3_-_42949526 | 3.87 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr5_-_57053687 | 3.73 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr8_-_19187191 | 3.65 |
|
|
|
| chr17_-_45021393 | 3.63 |
|
|
|
| chr23_-_43916621 | 3.62 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr1_+_43471683 | 3.61 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_+_16020464 | 3.58 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr11_-_25019899 | 3.56 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr11_-_6442588 | 3.56 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr2_-_37116119 | 3.49 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr3_-_42949616 | 3.43 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr14_-_33141111 | 3.41 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr20_-_34126039 | 3.38 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr15_-_19192862 | 3.37 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr5_+_3567992 | 3.35 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr12_+_30673985 | 3.34 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr17_+_14957568 | 3.32 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr22_-_22139268 | 3.30 |
ENSDART00000101659
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr12_-_33257026 | 3.29 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr11_+_24076334 | 3.29 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr24_-_9857510 | 3.29 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr21_+_10609580 | 3.28 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr3_-_26052601 | 3.26 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_-_31796734 | 3.22 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr6_-_42390995 | 3.21 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr5_+_65453249 | 3.14 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr12_-_33256754 | 3.13 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_+_67157818 | 3.13 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr17_+_31205274 | 3.12 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr20_-_34894930 | 3.05 |
|
|
|
| chr10_-_11011134 | 3.03 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr14_-_17270022 | 2.99 |
ENSDART00000123145
|
rnf4
|
ring finger protein 4 |
| KN150334v1_-_9585 | 2.97 |
ENSDART00000175935
|
CABZ01113810.1
|
ENSDARG00000107898 |
| chr2_-_45810787 | 2.96 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr8_-_2557556 | 2.94 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr6_-_9046536 | 2.93 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr14_+_29932533 | 2.93 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr17_+_22291546 | 2.92 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr3_-_39117855 | 2.92 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| chr13_-_37340209 | 2.92 |
|
|
|
| chr7_-_14149889 | 2.91 |
ENSDART00000166519
|
kif7
|
kinesin family member 7 |
| chr12_-_34657387 | 2.81 |
ENSDART00000153418
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr3_+_44928323 | 2.80 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr19_-_20578899 | 2.79 |
ENSDART00000151646
|
fam221a
|
family with sequence similarity 221, member A |
| chr12_-_33256934 | 2.79 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_+_25313170 | 2.77 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr5_-_28931727 | 2.76 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr2_+_23383492 | 2.75 |
ENSDART00000170669
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr5_-_29818204 | 2.72 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr22_+_21491685 | 2.71 |
ENSDART00000136374
|
mier2
|
mesoderm induction early response 1, family member 2 |
| chr14_+_14850200 | 2.69 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr13_+_36459562 | 2.66 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr17_-_25313024 | 2.64 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr15_+_29091983 | 2.58 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr17_+_24300451 | 2.58 |
ENSDART00000064083
|
otx1b
|
orthodenticle homeobox 1b |
| chr23_-_33783345 | 2.58 |
ENSDART00000143333
|
pou6f1
|
POU class 6 homeobox 1 |
| chr14_-_30642610 | 2.56 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr12_-_48759104 | 2.54 |
ENSDART00000130190
ENSDART00000105309 |
uros
|
uroporphyrinogen III synthase |
| chr23_+_43916520 | 2.53 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr17_-_43725091 | 2.53 |
|
|
|
| chr8_+_13327234 | 2.52 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr1_+_46474561 | 2.50 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr8_+_41003546 | 2.49 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr20_+_37023072 | 2.47 |
ENSDART00000155058
|
CR388421.1
|
ENSDARG00000096706 |
| chr8_-_26703757 | 2.45 |
ENSDART00000148033
|
kazna
|
kazrin, periplakin interacting protein a |
| chr21_-_20291586 | 2.43 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr22_+_2735606 | 2.41 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr17_-_12467563 | 2.38 |
|
|
|
| chr16_-_25452949 | 2.37 |
ENSDART00000167574
|
rbfa
|
ribosome binding factor A |
| KN150335v1_-_42498 | 2.36 |
|
|
|
| chr1_-_11191824 | 2.36 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr3_+_48811662 | 2.35 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr1_+_22868620 | 2.33 |
ENSDART00000133659
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr2_-_26941084 | 2.32 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_-_44924505 | 2.31 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr3_+_48811632 | 2.31 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr16_+_7463067 | 2.29 |
ENSDART00000149086
|
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr5_+_9579430 | 2.29 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr13_-_26668719 | 2.29 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr5_-_35997345 | 2.28 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr8_+_9660595 | 2.27 |
ENSDART00000091848
|
gripap1
|
GRIP1 associated protein 1 |
| chr23_+_22858063 | 2.27 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr14_-_26127173 | 2.27 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr21_-_22320879 | 2.27 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr2_+_19514886 | 2.26 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr24_-_24999240 | 2.25 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr5_-_29818325 | 2.25 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr5_-_4483970 | 2.24 |
|
|
|
| chr21_+_21242470 | 2.23 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr17_-_18777398 | 2.23 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr8_+_20108592 | 2.22 |
|
|
|
| chr10_+_20435265 | 2.22 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr6_-_46740359 | 2.19 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr24_-_30929990 | 2.18 |
|
|
|
| chr1_-_13302923 | 2.18 |
ENSDART00000011414
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr18_+_9755131 | 2.16 |
|
|
|
| chr3_+_44928592 | 2.15 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr4_-_20456437 | 2.15 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr21_-_32748287 | 2.12 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr19_+_39689450 | 2.12 |
|
|
|
| chr10_+_45184723 | 2.11 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr3_-_30754236 | 2.10 |
ENSDART00000109104
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr6_+_58628589 | 2.10 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr2_+_9854294 | 2.10 |
|
|
|
| chr14_+_38510523 | 2.10 |
|
|
|
| chr17_+_15780112 | 2.09 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr8_-_26703841 | 2.09 |
ENSDART00000148033
|
kazna
|
kazrin, periplakin interacting protein a |
| chr15_+_19389071 | 2.09 |
ENSDART00000164184
|
vps26b
|
VPS26 retromer complex component B |
| chr3_+_39437497 | 2.07 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_-_9794176 | 2.06 |
|
|
|
| chr19_-_20578863 | 2.05 |
ENSDART00000148179
|
fam221a
|
family with sequence similarity 221, member A |
| chr14_-_30642549 | 2.05 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr9_+_22166405 | 2.05 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr11_+_24076371 | 2.05 |
ENSDART00000166045
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr19_-_7114660 | 2.04 |
ENSDART00000124094
|
daxx
|
death-domain associated protein |
| chr22_-_10367866 | 2.04 |
ENSDART00000142886
|
nisch
|
nischarin |
| chr8_-_22493608 | 2.02 |
ENSDART00000021514
|
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr13_-_28480365 | 2.01 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr8_-_39850462 | 2.01 |
ENSDART00000131372
|
mlec
|
malectin |
| chr21_-_32747544 | 2.00 |
|
|
|
| chr23_+_43916552 | 1.99 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr8_-_1601007 | 1.99 |
ENSDART00000159705
|
dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr13_+_36459625 | 1.98 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr5_-_35997304 | 1.97 |
ENSDART00000031270
|
rhogc
|
ras homolog gene family, member Gc |
| chr3_+_60299938 | 1.97 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr19_+_43090603 | 1.95 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr3_-_42949449 | 1.95 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr10_-_3176372 | 1.95 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr2_-_26941232 | 1.92 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr23_+_22857958 | 1.92 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr7_-_38363533 | 1.92 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr15_+_22510807 | 1.92 |
ENSDART00000040542
|
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr4_-_20119812 | 1.90 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr19_+_7630777 | 1.90 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr14_+_14536088 | 1.88 |
ENSDART00000158291
|
slbp
|
stem-loop binding protein |
| chr3_+_35366065 | 1.86 |
ENSDART00000113219
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr24_-_20225190 | 1.85 |
ENSDART00000109223
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr10_+_15382807 | 1.85 |
ENSDART00000168909
|
trappc13
|
trafficking protein particle complex 13 |
| chr21_+_10609341 | 1.84 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr10_-_44173041 | 1.84 |
|
|
|
| chr3_+_1054368 | 1.84 |
ENSDART00000153693
|
si:ch73-166o21.1
|
si:ch73-166o21.1 |
| chr5_+_60028657 | 1.83 |
|
|
|
| chr16_+_42567707 | 1.81 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr3_-_32741894 | 1.81 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr21_-_2273151 | 1.78 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr8_-_2557506 | 1.77 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr14_+_38510427 | 1.76 |
|
|
|
| chr17_+_15780156 | 1.76 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr22_+_3989571 | 1.76 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr2_+_10349452 | 1.76 |
ENSDART00000153678
|
pfn2l
|
profilin 2 like |
| chr3_-_42949479 | 1.76 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr3_+_35292809 | 1.75 |
ENSDART00000176673
|
BX255942.1
|
ENSDARG00000108159 |
| chr12_+_5067663 | 1.75 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr16_-_17678748 | 1.73 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr7_+_20651600 | 1.73 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr16_-_43061368 | 1.72 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr14_+_23673635 | 1.72 |
ENSDART00000124944
|
kif3a
|
kinesin family member 3A |
| chr4_-_14916416 | 1.71 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr3_-_32205961 | 1.71 |
ENSDART00000156551
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr19_+_19806658 | 1.70 |
ENSDART00000160444
|
si:dkey-95j14.1
|
si:dkey-95j14.1 |
| chr6_+_39186673 | 1.70 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr22_-_16351654 | 1.69 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr3_+_39437613 | 1.69 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr12_+_47729417 | 1.69 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr15_-_5636737 | 1.69 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr21_-_2273243 | 1.69 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr16_+_9509605 | 1.68 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr15_-_16241412 | 1.68 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr8_+_12913443 | 1.67 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr24_-_20946041 | 1.67 |
ENSDART00000140786
|
qtrtd1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr1_-_8565788 | 1.67 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr2_+_31959127 | 1.67 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr4_+_71390236 | 1.67 |
ENSDART00000174116
|
zgc:152938
|
zgc:152938 |
| chr6_-_41141242 | 1.67 |
ENSDART00000128723
ENSDART00000151055 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr17_+_17745119 | 1.66 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr2_+_34675356 | 1.66 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0030238 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 2.1 | 8.6 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 1.8 | 12.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.4 | 4.1 | GO:0071846 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 1.1 | 3.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 1.1 | 4.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.1 | 5.3 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.0 | 3.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 1.0 | 4.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.0 | 2.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) temperature homeostasis(GO:0001659) |
| 0.9 | 3.8 | GO:0015809 | arginine transport(GO:0015809) |
| 0.9 | 2.7 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.8 | 6.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.7 | 2.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.7 | 2.1 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.7 | 3.4 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.7 | 3.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.6 | 2.5 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.6 | 2.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 2.4 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.6 | 2.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 3.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.6 | 5.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.6 | 2.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.6 | 1.7 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.5 | 5.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.5 | 1.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.5 | 1.4 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.5 | 3.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.5 | 1.8 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.5 | 2.7 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.5 | 4.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.4 | 1.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.4 | 1.7 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.4 | 1.7 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.4 | 1.7 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 0.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.4 | 1.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 1.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 3.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 1.3 | GO:1903650 | negative regulation of cytoplasmic transport(GO:1903650) |
| 0.3 | 3.2 | GO:0031204 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.6 | GO:0010518 | positive regulation of phospholipase activity(GO:0010518) positive regulation of lipase activity(GO:0060193) |
| 0.3 | 1.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 0.9 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.3 | 1.3 | GO:0046078 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.3 | 1.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.5 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 2.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.3 | 1.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 3.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.3 | 0.9 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.3 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 0.8 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 7.9 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.3 | 1.9 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 0.8 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.3 | 0.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 1.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.0 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 1.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.2 | 0.7 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 1.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.7 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 1.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 2.0 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 4.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 0.7 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.2 | 11.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.2 | 4.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 0.8 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 3.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 1.7 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 3.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 7.3 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.2 | 0.7 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.2 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 4.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.5 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 2.5 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 2.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.5 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 2.6 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 4.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 | 0.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.8 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 3.8 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 0.4 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.1 | 2.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 2.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 3.1 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.4 | GO:0039531 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.1 | 2.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 3.6 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 2.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.0 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 7.1 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.4 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 0.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.3 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.6 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 1.3 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.9 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 2.8 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 4.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 2.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.7 | GO:0010138 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 4.7 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.1 | 0.8 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 2.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.1 | GO:0002821 | positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) |
| 0.1 | 2.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 3.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.5 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 1.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.0 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 0.6 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 2.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 5.1 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.1 | 1.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 6.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.1 | 1.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 2.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.2 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.3 | GO:0007034 | vacuolar transport(GO:0007034) |
| 0.0 | 7.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.8 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.3 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.8 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 1.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.3 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.7 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 3.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.8 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 3.6 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.4 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.3 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 6.7 | GO:0006644 | phospholipid metabolic process(GO:0006644) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.8 | GO:0038127 | ERBB signaling pathway(GO:0038127) |
| 0.0 | 1.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.2 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.7 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 1.1 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 1.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 5.1 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.4 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 1.2 | 12.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.2 | 4.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 1.1 | 3.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 1.1 | 4.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 1.0 | 7.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.7 | 3.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.7 | 2.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 6.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 2.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.5 | 1.8 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.5 | 3.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.4 | 1.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.4 | 4.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 2.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 2.3 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.3 | 2.1 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 1.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.7 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 2.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 3.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 2.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 0.7 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.2 | 1.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 4.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 4.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.0 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.8 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.2 | 1.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 2.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 2.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 0.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 2.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 7.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 4.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 2.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 4.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.6 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 6.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 7.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.6 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 5.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 4.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 4.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 2.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 4.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.2 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 2.8 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 4.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 5.1 | GO:0031966 | mitochondrial envelope(GO:0005740) mitochondrial membrane(GO:0031966) |
| 0.0 | 6.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.3 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.3 | 9.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.3 | 9.1 | GO:0005537 | mannose binding(GO:0005537) |
| 1.0 | 4.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.9 | 2.8 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.7 | 2.9 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.7 | 2.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.7 | 2.7 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.7 | 2.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.7 | 2.0 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.6 | 2.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.6 | 3.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.6 | 1.7 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.6 | 2.8 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.5 | 4.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.5 | 5.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.5 | 3.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 1.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 2.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.4 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 1.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 1.0 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.3 | 2.0 | GO:0015562 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 1.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 5.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 1.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 3.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 2.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.3 | 7.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 1.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 1.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 1.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 2.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 2.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 3.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 9.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 3.2 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.2 | 1.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.3 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.4 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.2 | 0.9 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 2.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 1.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.8 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 2.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 6.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.5 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 3.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 2.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.5 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.8 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 3.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 2.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 2.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.3 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.1 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 3.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.8 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 2.2 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 3.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 2.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 4.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 5.8 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.6 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 6.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 10.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 3.1 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.4 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 3.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 5.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 11.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 6.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 1.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 4.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 2.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 1.0 | 9.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.8 | 5.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.7 | 14.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.6 | 4.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.5 | 6.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 1.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 3.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 1.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.7 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 2.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 5.9 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 5.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 3.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.4 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 3.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 2.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.8 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |