Project
DANIO-CODE
Navigation
Downloads
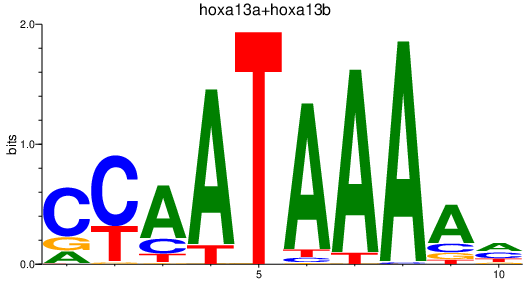
Results for hoxa13a+hoxa13b
Z-value: 0.93
Transcription factors associated with hoxa13a+hoxa13b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa13b
|
ENSDARG00000036254 | homeobox A13b |
|
hoxa13a
|
ENSDARG00000100312 | homeobox A13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa13a | dr10_dc_chr19_+_20156598_20156642 | -0.59 | 1.7e-02 | Click! |
| hoxa13b | dr10_dc_chr16_+_21090083_21090085 | -0.35 | 1.8e-01 | Click! |
Activity profile of hoxa13a+hoxa13b motif
Sorted Z-values of hoxa13a+hoxa13b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa13a+hoxa13b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_21587697 | 1.14 |
ENSDART00000029122
|
zgc:165539
|
zgc:165539 |
| chr6_+_46430090 | 1.12 |
ENSDART00000064865
ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr14_+_34154895 | 1.08 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr21_+_20360180 | 1.08 |
ENSDART00000003299
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr24_+_21495522 | 1.07 |
ENSDART00000105925
|
gtf3ab
|
general transcription factor IIIA, b |
| chr20_+_32598608 | 1.07 |
ENSDART00000021035
ENSDART00000152944 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr13_-_50299643 | 1.04 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr18_-_40718244 | 1.01 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr13_-_50299821 | 1.01 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr5_+_68036497 | 0.99 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr6_+_37323636 | 0.97 |
ENSDART00000178969
ENSDART00000104180 |
zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr11_+_44002664 | 0.95 |
|
|
|
| chr1_-_32914487 | 0.93 |
ENSDART00000015547
|
cldng
|
claudin g |
| chr11_+_7570027 | 0.91 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr19_+_14592632 | 0.91 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr11_+_7570097 | 0.90 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr21_+_17509116 | 0.90 |
|
|
|
| chr8_-_337744 | 0.89 |
|
|
|
| chr20_+_42640466 | 0.85 |
ENSDART00000134066
|
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr2_+_10349452 | 0.83 |
ENSDART00000153678
|
pfn2l
|
profilin 2 like |
| chr1_+_54005969 | 0.79 |
ENSDART00000077578
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr14_+_38509671 | 0.78 |
|
|
|
| chr14_+_22201617 | 0.75 |
ENSDART00000174265
|
smyd5
|
SMYD family member 5 |
| chr22_-_22139268 | 0.75 |
ENSDART00000101659
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr2_-_50491234 | 0.72 |
ENSDART00000165678
|
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr21_-_7322856 | 0.72 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| chr7_-_14149889 | 0.71 |
ENSDART00000166519
|
kif7
|
kinesin family member 7 |
| chr11_+_25302072 | 0.71 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr3_+_24004141 | 0.70 |
ENSDART00000155216
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr17_-_30884905 | 0.69 |
ENSDART00000131633
ENSDART00000146824 |
evla
|
Enah/Vasp-like a |
| chr25_+_7359375 | 0.69 |
|
|
|
| chr19_-_868378 | 0.68 |
|
|
|
| chr1_+_44516337 | 0.67 |
ENSDART00000125037
|
EVI5L
|
ecotropic viral integration site 5 like |
| chr16_+_29574449 | 0.66 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr11_-_25224150 | 0.66 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr12_+_19077980 | 0.66 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| KN150712v1_-_26150 | 0.65 |
|
|
|
| chr22_-_22139099 | 0.65 |
|
|
|
| chr20_+_32598826 | 0.65 |
ENSDART00000021035
ENSDART00000152944 |
sec63
|
SEC63 homolog, protein translocation regulator |
| chr21_-_45647945 | 0.65 |
|
|
|
| chr9_-_30997261 | 0.64 |
ENSDART00000146300
|
klf5b
|
Kruppel-like factor 5b |
| chr2_+_50680126 | 0.64 |
ENSDART00000122716
|
ENSDARG00000090398
|
ENSDARG00000090398 |
| chr14_+_38510523 | 0.63 |
|
|
|
| chr10_+_32122607 | 0.63 |
ENSDART00000062311
ENSDART00000124166 |
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr11_+_24076334 | 0.62 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr14_-_26127173 | 0.62 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr21_-_20291586 | 0.60 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr11_+_25302044 | 0.60 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr9_-_54126121 | 0.59 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr3_+_35140906 | 0.59 |
|
|
|
| chr13_+_50990946 | 0.58 |
|
|
|
| chr11_+_25302180 | 0.58 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr19_-_24971633 | 0.58 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr13_-_2296283 | 0.57 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr4_+_67053031 | 0.57 |
ENSDART00000158019
|
CR759907.2
|
ENSDARG00000099855 |
| chr16_-_55211053 | 0.56 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr18_+_20044862 | 0.56 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr2_+_11094785 | 0.55 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr4_-_16887296 | 0.55 |
ENSDART00000044005
ENSDART00000042874 ENSDART00000016690 |
tmpoa
|
thymopoietin a |
| chr14_+_29933552 | 0.55 |
|
|
|
| chr14_+_34150130 | 0.54 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr12_-_10474942 | 0.54 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr14_-_33141111 | 0.54 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr11_+_19440815 | 0.54 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr1_-_39141284 | 0.53 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr10_-_11011134 | 0.53 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr6_+_43452686 | 0.53 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr11_+_24076576 | 0.53 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_+_19610019 | 0.52 |
|
|
|
| chr4_-_71770315 | 0.52 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr7_+_67206056 | 0.52 |
ENSDART00000162553
|
kars
|
lysyl-tRNA synthetase |
| chr25_+_20596490 | 0.52 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr24_-_42037243 | 0.52 |
|
|
|
| chr5_+_67157818 | 0.51 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr22_-_20963723 | 0.51 |
|
|
|
| chr24_-_8592157 | 0.51 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr21_+_11656401 | 0.51 |
ENSDART00000102404
|
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr6_+_28218420 | 0.51 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr5_+_19950164 | 0.50 |
ENSDART00000008402
|
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr11_+_24076371 | 0.49 |
ENSDART00000166045
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr6_+_16342020 | 0.49 |
ENSDART00000114667
|
zgc:161969
|
zgc:161969 |
| chr19_-_14329389 | 0.49 |
ENSDART00000164594
|
ta
|
T, brachyury homolog a |
| chr9_+_34510091 | 0.49 |
ENSDART00000035522
ENSDART00000170615 ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr22_+_10752209 | 0.49 |
ENSDART00000081183
|
enc3
|
ectodermal-neural cortex 3 |
| chr2_-_6545431 | 0.49 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr23_-_39253291 | 0.49 |
|
|
|
| chr12_+_46242558 | 0.49 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr2_+_30198819 | 0.48 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr10_+_39142185 | 0.48 |
|
|
|
| chr23_+_7445760 | 0.48 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr18_+_20045293 | 0.48 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr21_-_2273151 | 0.48 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr11_+_36117256 | 0.48 |
ENSDART00000141529
|
atxn7l2a
|
ataxin 7-like 2a |
| chr21_-_3548863 | 0.48 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr25_+_7359012 | 0.47 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr7_+_15479700 | 0.47 |
|
|
|
| chr10_+_17919692 | 0.47 |
|
|
|
| chr14_-_23813378 | 0.47 |
ENSDART00000133522
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr8_-_18869085 | 0.47 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr15_-_5913264 | 0.46 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr17_-_49381216 | 0.46 |
ENSDART00000166394
|
fcf1
|
FCF1 rRNA-processing protein |
| chr5_-_30114979 | 0.46 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr5_-_30114539 | 0.46 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr1_-_8565788 | 0.46 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr7_-_25805221 | 0.46 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr5_-_32680882 | 0.46 |
ENSDART00000138116
|
surf6
|
surfeit 6 |
| chr13_+_17564330 | 0.45 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr12_+_27444732 | 0.45 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr14_+_22159893 | 0.44 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr18_+_20045221 | 0.44 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr1_-_25970826 | 0.44 |
ENSDART00000165460
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr5_-_36348618 | 0.44 |
|
|
|
| chr18_-_8422329 | 0.44 |
ENSDART00000081143
|
sephs1
|
selenophosphate synthetase 1 |
| chr22_+_15317622 | 0.44 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr7_+_34025638 | 0.43 |
ENSDART00000052471
ENSDART00000173798 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr2_+_10349152 | 0.43 |
ENSDART00000153678
|
pfn2l
|
profilin 2 like |
| chr16_+_43236190 | 0.43 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr7_+_5843895 | 0.43 |
ENSDART00000115062
|
CU459186.4
|
Histone H3.2 |
| chr4_-_14193396 | 0.43 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr5_-_30114910 | 0.43 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr5_-_25791559 | 0.43 |
|
|
|
| chr5_-_40503137 | 0.43 |
ENSDART00000051070
|
golph3
|
golgi phosphoprotein 3 |
| chr5_+_67157870 | 0.43 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr5_+_56658431 | 0.42 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr8_-_2557556 | 0.42 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr6_+_38882608 | 0.42 |
ENSDART00000019939
|
bin2b
|
bridging integrator 2b |
| chr6_+_2040661 | 0.42 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor, type IBb |
| chr14_+_38510427 | 0.42 |
|
|
|
| chr2_+_19546837 | 0.42 |
ENSDART00000163137
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr4_+_295319 | 0.41 |
ENSDART00000067489
|
si:ch211-51e12.7
|
si:ch211-51e12.7 |
| chr8_+_42910826 | 0.41 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr12_+_22552867 | 0.41 |
ENSDART00000152930
|
cdca9
|
cell division cycle associated 9 |
| chr11_+_42265857 | 0.41 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr15_-_5810906 | 0.41 |
ENSDART00000142334
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr14_-_32525979 | 0.41 |
ENSDART00000166133
|
ube2a
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog) |
| chr3_-_47825755 | 0.41 |
ENSDART00000145937
|
h3f3a
|
H3 histone, family 3A |
| chr25_+_7358777 | 0.41 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr8_-_2557506 | 0.41 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr2_-_11094083 | 0.40 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr7_+_51530818 | 0.40 |
ENSDART00000026571
|
slc38a7
|
solute carrier family 38, member 7 |
| chr18_+_46153484 | 0.40 |
ENSDART00000015034
|
blvrb
|
biliverdin reductase B |
| chr20_-_6153384 | 0.40 |
ENSDART00000161566
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr21_-_20291707 | 0.40 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr19_+_7916050 | 0.39 |
ENSDART00000026276
|
cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr12_-_27032151 | 0.39 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr21_-_36338514 | 0.39 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr10_-_6453139 | 0.39 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr6_+_27523853 | 0.39 |
ENSDART00000128985
|
ryk
|
receptor-like tyrosine kinase |
| chr23_+_22857958 | 0.39 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr5_-_28931727 | 0.39 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr10_+_30483855 | 0.38 |
|
|
|
| chr22_+_1837102 | 0.38 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr14_+_7585289 | 0.38 |
ENSDART00000161307
|
ube2d2
|
ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) |
| chr5_-_30115110 | 0.38 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr8_+_23840165 | 0.38 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr18_+_41559512 | 0.37 |
ENSDART00000059135
|
bcl7bb
|
B-cell CLL/lymphoma 7B, b |
| chr6_+_23709605 | 0.37 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr3_+_30962542 | 0.37 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr8_+_36951655 | 0.37 |
|
|
|
| chr5_+_40543392 | 0.37 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr2_-_45810787 | 0.37 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr8_-_20882626 | 0.37 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr7_-_25805806 | 0.37 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr23_+_43916552 | 0.37 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr19_-_4206333 | 0.37 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr15_-_5912917 | 0.37 |
|
|
|
| chr19_-_869177 | 0.37 |
|
|
|
| chr16_+_27608837 | 0.37 |
ENSDART00000147611
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr25_-_12810230 | 0.36 |
ENSDART00000165156
|
sept15
|
septin 15 |
| chr1_-_39265484 | 0.36 |
ENSDART00000160066
|
ing2
|
inhibitor of growth family, member 2 |
| chr23_-_43916621 | 0.36 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr23_-_27318468 | 0.36 |
|
|
|
| chr22_-_36718332 | 0.36 |
|
|
|
| chr14_+_29932533 | 0.36 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr23_+_22858063 | 0.36 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr3_-_39117855 | 0.35 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| chr6_+_46429927 | 0.35 |
ENSDART00000064865
ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr22_+_2329458 | 0.35 |
ENSDART00000143687
|
zgc:174224
|
zgc:174224 |
| chr4_-_7721419 | 0.35 |
|
|
|
| chr12_+_27444762 | 0.35 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr23_+_22858361 | 0.35 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr12_+_25341263 | 0.34 |
ENSDART00000153333
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr12_+_26943812 | 0.34 |
ENSDART00000153054
|
fbrs
|
fibrosin |
| chr25_+_3169073 | 0.34 |
|
|
|
| chr19_+_21209328 | 0.34 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr5_-_40503363 | 0.34 |
ENSDART00000074781
|
golph3
|
golgi phosphoprotein 3 |
| chr16_-_33047066 | 0.34 |
ENSDART00000147941
ENSDART00000075218 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr4_-_14916416 | 0.34 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr16_-_7876036 | 0.34 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr21_-_30371923 | 0.34 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr7_+_5835252 | 0.34 |
ENSDART00000170763
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr20_+_23707789 | 0.34 |
|
|
|
| chr9_+_2336680 | 0.34 |
ENSDART00000016417
|
atp5g3a
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9), genome duplicate a |
| chr18_+_20045506 | 0.34 |
|
|
|
| chr23_-_27318392 | 0.33 |
|
|
|
| chr7_+_53143890 | 0.33 |
ENSDART00000164643
|
ENSDARG00000104530
|
ENSDARG00000104530 |
| chr12_-_36415669 | 0.33 |
|
|
|
| chr12_-_11219985 | 0.33 |
ENSDART00000149229
|
si:ch73-30l9.1
|
si:ch73-30l9.1 |
| chr24_+_31751950 | 0.33 |
ENSDART00000156493
|
CT737162.1
|
ENSDARG00000096998 |
| chr3_+_35277555 | 0.33 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.6 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 1.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.8 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 2.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 0.7 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.2 | 1.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 1.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.5 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.2 | 1.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.5 | GO:0006226 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.5 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.3 | GO:0043393 | regulation of receptor recycling(GO:0001919) G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.1 | 0.5 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 1.0 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.1 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.3 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.5 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.3 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) negative regulation of skeletal muscle cell proliferation(GO:0014859) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.3 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.4 | GO:0030237 | female sex determination(GO:0030237) |
| 0.1 | 0.2 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0009070 | serine family amino acid biosynthetic process(GO:0009070) |
| 0.1 | 0.2 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 1.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.5 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.2 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.2 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 1.1 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.2 | GO:0071846 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 0.1 | 0.2 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.2 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) positive regulation of lipid transport(GO:0032370) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 1.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.7 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.4 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 0.5 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.2 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 1.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:1904035 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.3 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.6 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.2 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0034121 | regulation of toll-like receptor signaling pathway(GO:0034121) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.3 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 1.2 | GO:0007281 | germ cell development(GO:0007281) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.6 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 3.1 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0031207 | endoplasmic reticulum Sec complex(GO:0031205) Sec62/Sec63 complex(GO:0031207) |
| 0.4 | 1.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 0.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.6 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.4 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.2 | GO:0017054 | negative cofactor 2 complex(GO:0017054) |
| 0.1 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 1.0 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.6 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0032593 | lateral plasma membrane(GO:0016328) insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.0 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.9 | GO:0000793 | condensed chromosome(GO:0000793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0030882 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.7 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.2 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 0.8 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.2 | 0.7 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.7 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 0.5 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 0.7 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.2 | 0.5 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.1 | 0.3 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.3 | GO:0004352 | glutamate dehydrogenase (NAD+) activity(GO:0004352) glutamate dehydrogenase [NAD(P)+] activity(GO:0004353) oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.4 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.7 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.2 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.2 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.1 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.2 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.3 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 1.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.1 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |