Project
DANIO-CODE
Navigation
Downloads
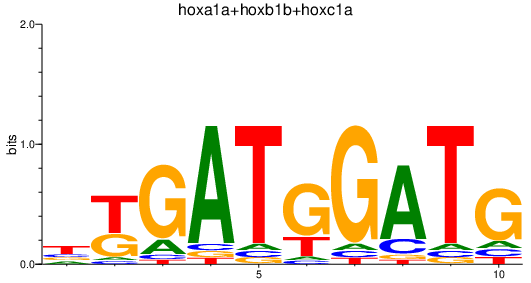
Results for hoxa1a+hoxb1b+hoxc1a
Z-value: 1.66
Transcription factors associated with hoxa1a+hoxb1b+hoxc1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb1b
|
ENSDARG00000054033 | homeobox B1b |
|
hoxc1a
|
ENSDARG00000070337 | homeobox C1a |
|
hoxa1a
|
ENSDARG00000104307 | homeobox A1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc1a | dr10_dc_chr23_+_36079624_36079679 | 0.98 | 5.5e-12 | Click! |
| hoxa1a | dr10_dc_chr19_+_20213125_20213150 | 0.91 | 7.7e-07 | Click! |
| hoxb1b | dr10_dc_chr12_+_27050043_27050092 | 0.72 | 1.5e-03 | Click! |
Activity profile of hoxa1a+hoxb1b+hoxc1a motif
Sorted Z-values of hoxa1a+hoxb1b+hoxc1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa1a+hoxb1b+hoxc1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_24932972 | 7.91 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr3_-_54893038 | 6.26 |
ENSDART00000155871
ENSDART00000109016 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr17_+_17841882 | 6.00 |
ENSDART00000123311
|
ism2a
|
isthmin 2a |
| chr5_-_65349550 | 5.26 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr20_-_9474672 | 5.24 |
ENSDART00000152674
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr6_+_60190849 | 5.12 |
|
|
|
| chr24_+_389982 | 4.89 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr21_-_43083936 | 4.70 |
ENSDART00000040169
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr5_+_16669696 | 4.54 |
|
|
|
| chr12_-_25825072 | 4.40 |
|
|
|
| chr20_-_32543497 | 4.22 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr6_+_29800606 | 4.13 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr19_-_2399429 | 4.06 |
ENSDART00000043595
|
twist1a
|
twist family bHLH transcription factor 1a |
| chr20_-_29580624 | 4.05 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr25_+_20118286 | 3.73 |
ENSDART00000104297
|
tnnt2d
|
troponin T2d, cardiac |
| chr19_-_47994946 | 3.43 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr7_+_16610457 | 3.32 |
|
|
|
| chr21_-_26459113 | 3.28 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| chr7_+_16583234 | 3.24 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr8_-_40425193 | 3.12 |
ENSDART00000130798
|
myl7
|
myosin, light chain 7, regulatory |
| chr14_+_503643 | 3.11 |
|
|
|
| chr23_+_2391223 | 3.02 |
|
|
|
| chr23_-_12079877 | 2.96 |
ENSDART00000147956
|
ENSDARG00000076364
|
ENSDARG00000076364 |
| chr3_-_45779817 | 2.84 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr11_+_43559400 | 2.81 |
|
|
|
| chr17_-_52020689 | 2.80 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr6_+_30441419 | 2.64 |
|
|
|
| chr18_-_23889256 | 2.63 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_+_21521123 | 2.63 |
ENSDART00000112531
ENSDART00000100936 |
tmem88b
|
transmembrane protein 88 b |
| chr18_+_24935499 | 2.52 |
|
|
|
| chr21_+_16987306 | 2.49 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr20_-_35938683 | 2.41 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr16_+_46435014 | 2.41 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr16_+_14817799 | 2.38 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr3_-_44113070 | 2.37 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr23_+_36079164 | 2.36 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr5_+_19802818 | 2.31 |
|
|
|
| chr23_+_36079624 | 2.19 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr3_+_23638277 | 2.19 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr24_+_40974634 | 2.16 |
|
|
|
| chr16_+_10531986 | 2.12 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr3_+_15656123 | 2.03 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr7_+_42124857 | 2.02 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr16_+_46902372 | 1.99 |
ENSDART00000177679
|
thsd7ab
|
thrombospondin, type I, domain containing 7Ab |
| chr1_+_45503061 | 1.96 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr17_-_6915458 | 1.90 |
ENSDART00000081797
|
sash1b
|
SAM and SH3 domain containing 1b |
| chr9_+_32267615 | 1.85 |
|
|
|
| chr11_+_34562255 | 1.84 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
| chr17_+_22936598 | 1.78 |
ENSDART00000155145
|
ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr24_-_26744092 | 1.77 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr4_-_14927871 | 1.73 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr8_-_17692185 | 1.72 |
|
|
|
| chr18_-_23889025 | 1.70 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr10_+_17494274 | 1.68 |
|
|
|
| chr1_+_23093114 | 1.65 |
|
|
|
| chr18_-_23888988 | 1.62 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_-_26586506 | 1.61 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr20_+_27792255 | 1.60 |
ENSDART00000153299
|
si:dkey-1h6.8
|
si:dkey-1h6.8 |
| chr21_-_14534576 | 1.59 |
ENSDART00000089967
ENSDART00000132142 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr5_-_71295651 | 1.58 |
|
|
|
| chr9_-_23406315 | 1.57 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr7_+_73466424 | 1.55 |
ENSDART00000123429
|
si:dkey-46i9.6
|
si:dkey-46i9.6 |
| chr4_-_1564624 | 1.53 |
ENSDART00000168633
|
BICD1 (1 of many)
|
BICD cargo adaptor 1 |
| chr16_-_16796313 | 1.50 |
|
|
|
| chr23_-_2027166 | 1.49 |
ENSDART00000127443
|
prdm5
|
PR domain containing 5 |
| chr16_+_21121428 | 1.49 |
|
|
|
| chr23_-_19053587 | 1.45 |
|
|
|
| chr13_+_35619748 | 1.43 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr5_+_6054781 | 1.43 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr11_-_11487856 | 1.42 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr14_+_23917724 | 1.42 |
ENSDART00000138082
ENSDART00000079215 |
stc2a
|
stanniocalcin 2a |
| chr22_-_15576473 | 1.39 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr18_-_23889169 | 1.33 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_+_34207218 | 1.33 |
ENSDART00000135608
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr22_-_34961734 | 1.22 |
ENSDART00000165142
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr7_-_71929355 | 1.16 |
|
|
|
| chr10_-_27780050 | 1.15 |
ENSDART00000138149
|
si:dkey-33o22.1
|
si:dkey-33o22.1 |
| chr24_+_27469268 | 1.15 |
ENSDART00000105774
|
ek1
|
eph-like kinase 1 |
| chr3_-_55399331 | 1.14 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr20_+_22780756 | 1.11 |
|
|
|
| chr18_+_5707066 | 1.09 |
|
|
|
| chr22_-_562269 | 1.03 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr14_-_46645127 | 1.01 |
|
|
|
| chr17_-_19606453 | 0.99 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr1_-_25225339 | 0.99 |
ENSDART00000109714
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr1_+_19376127 | 0.97 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr17_-_8016214 | 0.96 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr9_+_32267297 | 0.95 |
|
|
|
| chr21_+_10699071 | 0.93 |
|
|
|
| chr5_-_51631013 | 0.92 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr1_-_20132900 | 0.90 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr14_-_26078923 | 0.90 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr4_-_5643440 | 0.89 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr3_-_60723276 | 0.84 |
|
|
|
| chr7_+_30803603 | 0.81 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr10_+_20151165 | 0.76 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr20_-_32543458 | 0.75 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr1_+_23093475 | 0.73 |
|
|
|
| chr16_-_31230568 | 0.73 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr2_-_16548451 | 0.73 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr3_-_40387858 | 0.72 |
ENSDART00000055194
|
actb2
|
actin, beta 2 |
| chr4_-_70713033 | 0.72 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr14_+_43599635 | 0.71 |
ENSDART00000155539
|
CR786577.1
|
ENSDARG00000097875 |
| chr7_+_29681510 | 0.69 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr24_+_34227202 | 0.69 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr7_+_16583304 | 0.67 |
ENSDART00000113332
|
nav2a
|
neuron navigator 2a |
| chr18_+_41552295 | 0.67 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr22_+_35113233 | 0.66 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr21_-_28304276 | 0.66 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr3_+_23621561 | 0.65 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr21_-_36886505 | 0.65 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr7_-_46510731 | 0.64 |
|
|
|
| chr13_-_43027245 | 0.62 |
ENSDART00000099601
|
ENSDARG00000068784
|
ENSDARG00000068784 |
| chr21_+_10698503 | 0.61 |
ENSDART00000144460
|
znf532
|
zinc finger protein 532 |
| chr17_-_44895892 | 0.58 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr20_+_29685096 | 0.57 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr5_+_60457842 | 0.54 |
|
|
|
| chr23_+_2391317 | 0.53 |
|
|
|
| chr12_+_16848206 | 0.49 |
|
|
|
| chr5_-_71277652 | 0.48 |
|
|
|
| chr14_+_4169371 | 0.45 |
ENSDART00000136665
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr3_+_60724858 | 0.45 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr25_+_7287952 | 0.44 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr5_+_8460247 | 0.42 |
ENSDART00000091397
|
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr17_-_44895849 | 0.41 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr16_-_29502678 | 0.41 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr17_+_44896297 | 0.37 |
ENSDART00000133103
|
coq6
|
coenzyme Q6 monooxygenase |
| chr20_-_53264360 | 0.37 |
ENSDART00000124231
|
ENSDARG00000090191
|
ENSDARG00000090191 |
| chr4_-_13256905 | 0.34 |
ENSDART00000026593
ENSDART00000150577 |
grip1
|
glutamate receptor interacting protein 1 |
| chr15_+_32139366 | 0.33 |
ENSDART00000153791
|
dchs1a
|
dachsous cadherin-related 1a |
| chr9_+_32262914 | 0.33 |
ENSDART00000110204
|
pikfyve
|
phosphoinositide kinase, FYVE finger containing |
| chr10_+_20151082 | 0.32 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| KN150266v1_-_69477 | 0.32 |
|
|
|
| chr14_+_18956828 | 0.32 |
ENSDART00000005738
|
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr4_-_12107094 | 0.28 |
ENSDART00000048675
|
mrps33
|
mitochondrial ribosomal protein S33 |
| chr8_+_18800414 | 0.27 |
ENSDART00000160732
|
mpnd
|
MPN domain containing |
| chr17_+_51675406 | 0.23 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr7_+_13130332 | 0.23 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr22_-_15576405 | 0.22 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr23_-_16765933 | 0.21 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr22_+_11827925 | 0.21 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr21_-_28304413 | 0.18 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr17_+_44896084 | 0.17 |
ENSDART00000085009
|
coq6
|
coenzyme Q6 monooxygenase |
| chr2_-_14833381 | 0.14 |
|
|
|
| chr12_-_30898994 | 0.13 |
|
|
|
| chr22_+_1121048 | 0.12 |
ENSDART00000158017
|
ENSDARG00000100747
|
ENSDARG00000100747 |
| chr6_-_31589470 | 0.08 |
ENSDART00000111837
|
raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr20_+_29685331 | 0.06 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr13_-_36164801 | 0.02 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr10_+_4717721 | 0.02 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 1.5 | 7.3 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 1.0 | 3.1 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.8 | 4.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.8 | 2.4 | GO:0050864 | regulation of B cell activation(GO:0050864) |
| 0.6 | 6.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.2 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.4 | 5.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.4 | 1.8 | GO:0015817 | histidine transport(GO:0015817) |
| 0.3 | 1.6 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.3 | 0.8 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 1.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.2 | 0.9 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 2.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 1.1 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.2 | 4.8 | GO:0030278 | regulation of ossification(GO:0030278) |
| 0.1 | 0.4 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 3.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 3.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 1.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 4.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.5 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 0.7 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 2.8 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.5 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.4 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 2.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 2.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.2 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 2.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 1.6 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.5 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 2.4 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.9 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 5.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 2.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.4 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.7 | GO:0006936 | muscle contraction(GO:0006936) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.3 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 3.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.5 | 8.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 4.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 0.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.2 | 3.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.1 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.8 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.0 | 4.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.9 | 6.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.9 | 3.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.9 | 8.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.7 | 7.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.5 | 3.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 1.8 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.3 | 2.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.4 | GO:0016493 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 2.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 0.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 2.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 5.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.7 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.1 | 1.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 6.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 6.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 16.9 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 1.0 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 7.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.4 | 8.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 5.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 7.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 2.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |