Project
DANIO-CODE
Navigation
Downloads
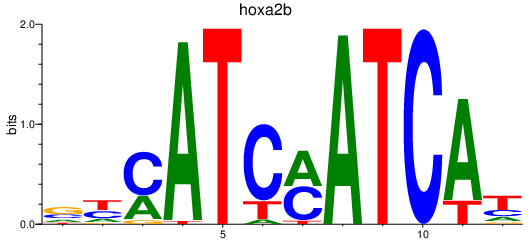
Results for hoxa2b
Z-value: 0.80
Transcription factors associated with hoxa2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa2b
|
ENSDARG00000023031 | homeobox A2b |
Activity profile of hoxa2b motif
Sorted Z-values of hoxa2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_26732762 | 4.45 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr8_+_21405821 | 3.65 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr8_+_21405788 | 2.26 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr6_-_53145464 | 1.82 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr21_-_43660946 | 1.77 |
ENSDART00000136392
|
si:ch211-263m18.4
|
si:ch211-263m18.4 |
| chr21_+_6890129 | 1.73 |
|
|
|
| chr19_-_27093095 | 1.69 |
|
|
|
| chr21_+_25740782 | 1.67 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr23_+_32174669 | 1.66 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr12_-_4264663 | 1.63 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| chr3_+_18786265 | 1.57 |
ENSDART00000141637
|
si:ch211-198m1.1
|
si:ch211-198m1.1 |
| chr2_+_28199458 | 1.49 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr11_+_23799984 | 1.47 |
|
|
|
| chr5_-_36503296 | 1.38 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr22_-_20670164 | 1.35 |
ENSDART00000169077
|
org
|
oogenesis-related gene |
| chr1_-_26321129 | 1.35 |
ENSDART00000147414
|
cntln
|
centlein, centrosomal protein |
| chr23_-_45107021 | 1.28 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr24_+_5782465 | 1.25 |
ENSDART00000178238
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr4_-_1522439 | 1.18 |
ENSDART00000175566
|
CABZ01085702.1
|
ENSDARG00000106927 |
| chr7_+_1337856 | 1.16 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr25_+_18855868 | 1.10 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr2_+_56534374 | 1.08 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr8_-_38322610 | 1.03 |
ENSDART00000134283
ENSDART00000132077 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr17_+_19606608 | 1.02 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr22_-_38321005 | 1.02 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr9_+_8387050 | 1.01 |
ENSDART00000136847
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr23_+_32175042 | 1.01 |
ENSDART00000138849
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr9_+_8412489 | 0.99 |
ENSDART00000144851
|
zgc:113984
|
zgc:113984 |
| chr24_+_19270877 | 0.98 |
|
|
|
| chr21_+_10609341 | 0.96 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr14_-_30578373 | 0.94 |
ENSDART00000176631
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr23_-_43785497 | 0.93 |
ENSDART00000165963
|
ENSDARG00000102050
|
ENSDARG00000102050 |
| chr6_-_53145335 | 0.93 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr18_+_18124107 | 0.91 |
ENSDART00000151406
|
BX649612.1
|
ENSDARG00000096293 |
| chr24_+_41991604 | 0.91 |
|
|
|
| chr23_+_43785682 | 0.90 |
ENSDART00000172160
|
ENSDARG00000105093
|
ENSDARG00000105093 |
| chr7_+_30355221 | 0.87 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr17_+_15666420 | 0.86 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr9_+_1820601 | 0.86 |
|
|
|
| chr9_-_55878791 | 0.85 |
|
|
|
| chr23_+_28396415 | 0.84 |
ENSDART00000142179
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr16_+_9509875 | 0.81 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| KN150623v1_+_258 | 0.81 |
|
|
|
| chr1_-_8593850 | 0.80 |
ENSDART00000146065
ENSDART00000114876 |
ubn1
|
ubinuclein 1 |
| chr7_-_24604255 | 0.79 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr10_+_7634671 | 0.79 |
ENSDART00000171744
|
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr10_-_7513764 | 0.78 |
ENSDART00000167054
ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr6_-_53145582 | 0.78 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr4_-_17289857 | 0.77 |
ENSDART00000178686
|
lrmp
|
lymphoid-restricted membrane protein |
| chr19_-_40111964 | 0.76 |
|
|
|
| chr11_-_18078147 | 0.76 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr6_-_2000017 | 0.76 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr5_+_29193876 | 0.75 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr24_-_41275160 | 0.74 |
|
|
|
| chr9_-_6861630 | 0.74 |
|
|
|
| chr25_+_20174490 | 0.74 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr8_+_5222023 | 0.74 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr18_-_25065264 | 0.73 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr14_+_32578253 | 0.73 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr23_-_29886117 | 0.73 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr16_-_29755471 | 0.70 |
ENSDART00000133965
|
scnm1
|
sodium channel modifier 1 |
| chr5_+_32324507 | 0.70 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr1_+_23685562 | 0.69 |
ENSDART00000102542
ENSDART00000160882 |
ENSDARG00000090822
qdprb2
|
ENSDARG00000090822 quinoid dihydropteridine reductase b2 |
| chr22_+_31073990 | 0.69 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr24_+_41991566 | 0.69 |
|
|
|
| chr22_-_10562118 | 0.68 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| KN150633v1_+_13413 | 0.68 |
|
|
|
| KN150339v1_-_39357 | 0.67 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr3_-_40090673 | 0.67 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr14_+_14850200 | 0.66 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr21_-_43660908 | 0.66 |
ENSDART00000136392
|
si:ch211-263m18.4
|
si:ch211-263m18.4 |
| chr3_-_20912934 | 0.66 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr1_+_46474561 | 0.66 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr5_-_11162837 | 0.66 |
|
|
|
| chr23_+_44814692 | 0.66 |
|
|
|
| chr18_+_17611571 | 0.66 |
ENSDART00000151850
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr17_+_28689850 | 0.65 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr14_+_25973961 | 0.65 |
ENSDART00000178140
ENSDART00000179471 |
FO082779.2
|
ENSDARG00000106651 |
| chr10_-_31861975 | 0.65 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr24_+_14792755 | 0.64 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr21_-_44086464 | 0.64 |
ENSDART00000130833
|
FO704810.1
|
ENSDARG00000087492 |
| chr12_+_48590478 | 0.64 |
|
|
|
| chr6_+_131451 | 0.64 |
ENSDART00000166361
|
fdx1l
|
ferredoxin 1-like |
| chr23_-_45107075 | 0.63 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr7_+_38044801 | 0.63 |
ENSDART00000160197
|
BX323855.1
|
ENSDARG00000098940 |
| chr4_+_13626978 | 0.63 |
ENSDART00000108928
|
pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr24_-_31967674 | 0.63 |
ENSDART00000156060
|
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr6_-_32424559 | 0.63 |
ENSDART00000151002
|
usp1
|
ubiquitin specific peptidase 1 |
| chr1_+_35253862 | 0.62 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr19_+_14132374 | 0.62 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr5_+_26888419 | 0.62 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr5_+_62611738 | 0.62 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr7_-_58856806 | 0.62 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr13_-_23626146 | 0.62 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr9_+_22846286 | 0.61 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr9_+_3577477 | 0.61 |
ENSDART00000147650
|
tlk1a
|
tousled-like kinase 1a |
| chr21_+_15774822 | 0.61 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr16_+_42567707 | 0.61 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr10_-_31862004 | 0.60 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr19_+_14132473 | 0.60 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr6_+_18894174 | 0.60 |
ENSDART00000165806
|
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr7_-_58856862 | 0.59 |
ENSDART00000164104
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr24_+_7855521 | 0.59 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr3_-_59690168 | 0.59 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr2_+_37312637 | 0.59 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr17_+_49537268 | 0.59 |
ENSDART00000123641
|
zgc:113372
|
zgc:113372 |
| chr6_-_53145759 | 0.59 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr14_-_36101782 | 0.59 |
|
|
|
| chr22_-_12738593 | 0.58 |
ENSDART00000143574
|
rqcd1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr25_-_12148502 | 0.58 |
|
|
|
| chr10_-_33435736 | 0.58 |
ENSDART00000023509
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_57056091 | 0.58 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr5_-_47419154 | 0.58 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr23_+_44814525 | 0.58 |
|
|
|
| chr5_+_64397858 | 0.58 |
ENSDART00000169209
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr2_-_15364526 | 0.58 |
ENSDART00000131835
|
hccsa.1
|
holocytochrome c synthase a |
| chr10_-_8088063 | 0.57 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr4_-_20514831 | 0.57 |
ENSDART00000146621
|
stk38l
|
serine/threonine kinase 38 like |
| chr10_+_42841562 | 0.57 |
|
|
|
| chr3_-_32982759 | 0.56 |
|
|
|
| chr16_+_9509747 | 0.56 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr8_-_50151276 | 0.56 |
|
|
|
| chr2_-_7913468 | 0.55 |
ENSDART00000163175
|
dcun1d1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| chr12_-_33481445 | 0.55 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr12_-_20494169 | 0.55 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr10_+_15382647 | 0.55 |
ENSDART00000046274
|
trappc13
|
trafficking protein particle complex 13 |
| chr16_+_41004372 | 0.55 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr5_+_62666247 | 0.55 |
ENSDART00000142862
|
ttf1
|
transcription termination factor, RNA polymerase I |
| chr5_+_29791993 | 0.54 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr8_-_38322559 | 0.54 |
ENSDART00000134283
ENSDART00000132077 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr25_+_7261050 | 0.54 |
ENSDART00000163017
|
prc1a
|
protein regulator of cytokinesis 1a |
| chr17_+_22050088 | 0.54 |
|
|
|
| chr7_-_19080002 | 0.54 |
ENSDART00000166121
ENSDART00000169668 |
dock11
|
dedicator of cytokinesis 11 |
| chr5_+_62666161 | 0.54 |
ENSDART00000142862
|
ttf1
|
transcription termination factor, RNA polymerase I |
| chr23_+_44814646 | 0.53 |
|
|
|
| KN149855v1_+_18816 | 0.53 |
|
|
|
| chr4_-_3340315 | 0.53 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr6_+_28001318 | 0.53 |
ENSDART00000143974
|
amotl2a
|
angiomotin like 2a |
| chr21_+_15774697 | 0.52 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr7_+_23938868 | 0.52 |
ENSDART00000168257
|
CR318603.1
|
ENSDARG00000099215 |
| chr3_-_6531056 | 0.51 |
ENSDART00000160979
|
jpt1b
|
Jupiter microtubule associated homolog 1b |
| chr2_-_6153363 | 0.51 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr4_-_1809690 | 0.51 |
|
|
|
| chr13_-_22921202 | 0.51 |
ENSDART00000111774
|
supv3l1
|
SUV3-like helicase |
| chr3_-_14545237 | 0.51 |
ENSDART00000133850
|
gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr5_-_14783255 | 0.51 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr17_-_12467563 | 0.50 |
|
|
|
| chr2_+_42077559 | 0.50 |
ENSDART00000023208
|
ENSDARG00000014209
|
ENSDARG00000014209 |
| chr3_+_14583287 | 0.50 |
|
|
|
| chr22_-_52569 | 0.50 |
|
|
|
| chr15_+_46053808 | 0.50 |
|
|
|
| chr2_-_38223813 | 0.50 |
ENSDART00000137395
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr18_+_5707331 | 0.49 |
|
|
|
| chr4_-_14916491 | 0.49 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr18_-_18886144 | 0.49 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr21_-_25358498 | 0.49 |
ENSDART00000167189
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr12_-_4264610 | 0.49 |
ENSDART00000152377
|
ca15b
|
carbonic anhydrase XVb |
| chr23_+_39227734 | 0.49 |
ENSDART00000065331
|
sall4
|
spalt-like transcription factor 4 |
| chr5_-_53777415 | 0.49 |
ENSDART00000169270
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr1_+_47323244 | 0.48 |
|
|
|
| chr2_+_46210909 | 0.48 |
|
|
|
| chr13_-_25277861 | 0.48 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr25_+_16784585 | 0.48 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr5_+_57056176 | 0.48 |
ENSDART00000134684
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr12_-_27121343 | 0.48 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr7_-_29084775 | 0.48 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr12_-_11419332 | 0.47 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr13_+_38688704 | 0.47 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr3_-_19890717 | 0.47 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr2_+_46210971 | 0.47 |
|
|
|
| chr11_-_18090243 | 0.47 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr5_+_35186037 | 0.47 |
ENSDART00000127383
ENSDART00000022043 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr25_+_18855775 | 0.46 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr15_-_25158683 | 0.46 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr19_-_44161905 | 0.46 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr20_-_14885599 | 0.46 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr16_+_29756032 | 0.46 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr19_+_20404995 | 0.46 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr13_-_30031075 | 0.46 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| KN150034v1_+_1223 | 0.46 |
|
|
|
| chr8_+_23083842 | 0.45 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr20_+_38354928 | 0.45 |
ENSDART00000149160
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr20_+_16274045 | 0.45 |
ENSDART00000129633
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr19_-_3833037 | 0.45 |
ENSDART00000168433
|
btr20
|
bloodthirsty-related gene family, member 20 |
| chr15_-_25158558 | 0.45 |
ENSDART00000142609
|
exo5
|
exonuclease 5 |
| chr25_-_34648876 | 0.45 |
ENSDART00000154851
|
zgc:153405
|
zgc:153405 |
| chr10_-_17593407 | 0.45 |
ENSDART00000145077
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr22_+_38928297 | 0.44 |
ENSDART00000133067
ENSDART00000085701 |
AL953867.2
senp5
|
ENSDARG00000095689 SUMO1/sentrin specific peptidase 5 |
| chr5_-_14000166 | 0.44 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr17_-_49381216 | 0.44 |
ENSDART00000166394
|
fcf1
|
FCF1 rRNA-processing protein |
| chr5_+_62628471 | 0.44 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr4_+_9357791 | 0.44 |
|
|
|
| chr6_+_28001359 | 0.44 |
ENSDART00000139367
|
amotl2a
|
angiomotin like 2a |
| chr5_+_62682097 | 0.44 |
ENSDART00000137855
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr23_+_35870640 | 0.44 |
ENSDART00000139057
|
BX465864.2
|
ENSDARG00000095755 |
| chr8_+_23637504 | 0.43 |
ENSDART00000083605
|
tbc1d25
|
TBC1 domain family, member 25 |
| chr21_+_6091802 | 0.43 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr19_+_47023218 | 0.43 |
|
|
|
| chr19_-_44161951 | 0.43 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr20_-_14885629 | 0.43 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.4 | 4.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 1.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 0.8 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.3 | 0.8 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.3 | 0.8 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 1.0 | GO:1902236 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.2 | 0.4 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.2 | 3.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 0.5 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.2 | 1.6 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.7 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.4 | GO:0090277 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.5 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.1 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.3 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.9 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.3 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 3.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.0 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.3 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.7 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.1 | 1.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.3 | GO:1990438 | snRNA 2'-O-methylation(GO:1990437) U6 2'-O-snRNA methylation(GO:1990438) |
| 0.1 | 0.3 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.5 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 1.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.7 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.7 | GO:1902686 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.3 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 1.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.4 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) regulation of mitotic attachment of spindle microtubules to kinetochore(GO:1902423) positive regulation of attachment of mitotic spindle microtubules to kinetochore(GO:1902425) RNA localization to chromatin(GO:1990280) |
| 0.1 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.5 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.7 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0046661 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.0 | 0.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.6 | GO:0051923 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) sulfation(GO:0051923) |
| 0.0 | 1.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.4 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.0 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.7 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 1.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.8 | GO:1903708 | positive regulation of hemopoiesis(GO:1903708) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.4 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.1 | GO:0042665 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.0 | 0.4 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.1 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.4 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 0.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.7 | GO:0031349 | positive regulation of defense response(GO:0031349) positive regulation of innate immune response(GO:0045089) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.7 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.4 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.5 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.0 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.5 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.1 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.4 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.7 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.3 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 1.9 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.5 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.4 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 3.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.4 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.3 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.6 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 2.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 5.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 1.1 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.3 | 3.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 0.9 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 1.6 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 1.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 4.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.0 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.3 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0008251 | tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.4 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 5.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 1.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0042923 | opioid receptor activity(GO:0004985) neuropeptide binding(GO:0042923) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 4.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.0 | 0.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |