Project
DANIO-CODE
Navigation
Downloads
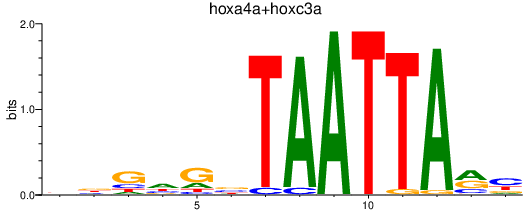
Results for hoxa4a+hoxc3a
Z-value: 0.65
Transcription factors associated with hoxa4a+hoxc3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc3a
|
ENSDARG00000070339 | homeobox C3a |
|
hoxa4a
|
ENSDARG00000103862 | homeobox A4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc3a | dr10_dc_chr23_+_35988426_35988442 | -0.17 | 5.2e-01 | Click! |
| hoxa4a | dr10_dc_chr19_+_20181908_20181912 | -0.10 | 7.0e-01 | Click! |
Activity profile of hoxa4a+hoxc3a motif
Sorted Z-values of hoxa4a+hoxc3a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa4a+hoxc3a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_32027717 | 0.92 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr11_-_1524107 | 0.90 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr1_-_54570813 | 0.89 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr18_-_40718244 | 0.82 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr23_+_28396415 | 0.80 |
ENSDART00000142179
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr21_+_25740782 | 0.73 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr16_+_42567707 | 0.68 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr15_+_34734212 | 0.53 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr10_-_35313462 | 0.53 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr20_-_37910887 | 0.53 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_+_36168475 | 0.51 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr6_+_28218420 | 0.51 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr10_+_33449922 | 0.50 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr9_+_45193290 | 0.50 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr5_+_39885307 | 0.50 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr19_-_1571878 | 0.49 |
|
|
|
| chr20_+_29306677 | 0.48 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr10_+_15296880 | 0.48 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr23_-_33692244 | 0.46 |
|
|
|
| chr10_-_21404605 | 0.44 |
ENSDART00000125167
|
avd
|
avidin |
| chr2_+_42923652 | 0.44 |
ENSDART00000168318
|
BX005083.1
|
ENSDARG00000097562 |
| chr19_-_4876806 | 0.43 |
ENSDART00000141336
ENSDART00000110551 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr11_-_6442588 | 0.43 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr22_-_20141589 | 0.43 |
ENSDART00000085913
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr18_+_8943793 | 0.42 |
ENSDART00000144247
|
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr19_-_5186692 | 0.41 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_-_20141216 | 0.41 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr22_-_864745 | 0.39 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr5_-_9320252 | 0.39 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr10_+_35313772 | 0.39 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr3_+_479366 | 0.38 |
|
|
|
| chr1_-_22617455 | 0.38 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr2_+_3115593 | 0.37 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr9_-_711269 | 0.37 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr6_+_7257078 | 0.37 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr10_+_2656840 | 0.37 |
|
|
|
| chr20_+_14218237 | 0.36 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr9_+_30610756 | 0.35 |
ENSDART00000145025
|
zgc:113314
|
zgc:113314 |
| chr14_+_24637864 | 0.35 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr7_+_15059782 | 0.34 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| KN150703v1_+_15225 | 0.34 |
|
|
|
| chr17_+_37280302 | 0.33 |
ENSDART00000128879
|
hadhab
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit b |
| chr2_+_11422195 | 0.33 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr2_-_26941084 | 0.32 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr13_-_30741976 | 0.32 |
ENSDART00000142535
|
CR762493.1
|
ENSDARG00000092411 |
| chr7_-_58863023 | 0.32 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr17_-_37447869 | 0.31 |
ENSDART00000148160
|
crip1
|
cysteine-rich protein 1 |
| chr9_-_30765708 | 0.31 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr23_+_2786407 | 0.31 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr5_+_25133592 | 0.31 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr20_-_16553968 | 0.30 |
ENSDART00000159053
|
gtf2a1
|
general transcription factor IIA, 1 |
| chr24_-_39997875 | 0.30 |
ENSDART00000143476
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr6_-_49900124 | 0.29 |
ENSDART00000150204
|
atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr3_-_19890717 | 0.29 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| KN149681v1_+_63726 | 0.29 |
ENSDART00000171794
|
apooa
|
apolipoprotein O, a |
| chr14_-_8634381 | 0.29 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr15_+_21327206 | 0.29 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr3_-_19890791 | 0.29 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr18_+_5066229 | 0.29 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr14_-_33141111 | 0.28 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr6_+_18941135 | 0.27 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr13_-_18564182 | 0.27 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr15_-_47260834 | 0.27 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr9_-_35824470 | 0.27 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr7_+_51499409 | 0.27 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr25_-_9889107 | 0.27 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr21_+_19040595 | 0.26 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr17_+_24595050 | 0.26 |
ENSDART00000064738
|
atpif1b
|
ATPase inhibitory factor 1b |
| chr10_+_44853207 | 0.26 |
ENSDART00000169466
|
scarb1
|
scavenger receptor class B, member 1 |
| chr16_+_42567668 | 0.26 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr12_+_22459177 | 0.26 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr25_-_19510386 | 0.26 |
ENSDART00000147223
|
gtse1
|
G-2 and S-phase expressed 1 |
| chr15_+_29207125 | 0.26 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr15_+_35089305 | 0.25 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr18_+_39506453 | 0.25 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr4_-_3340315 | 0.25 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr19_-_18664720 | 0.25 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| KN149947v1_-_2715 | 0.25 |
ENSDART00000158566
|
ENSDARG00000101294
|
ENSDARG00000101294 |
| chr11_+_44108192 | 0.25 |
ENSDART00000165219
ENSDART00000160678 |
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr21_+_27477153 | 0.25 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr11_-_44107913 | 0.24 |
|
|
|
| chr3_-_30948297 | 0.24 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr13_-_10214396 | 0.24 |
ENSDART00000132231
|
AL929457.1
|
ENSDARG00000095483 |
| chr20_+_29306945 | 0.24 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_+_36909452 | 0.23 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr5_+_1461715 | 0.23 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| KN150030v1_-_22572 | 0.23 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr18_-_3056732 | 0.23 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr5_-_3673698 | 0.22 |
|
|
|
| chr19_+_770458 | 0.22 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr25_-_13607319 | 0.22 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr24_+_17115897 | 0.22 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr2_-_36845638 | 0.22 |
|
|
|
| chr6_+_49902765 | 0.22 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr1_+_50547341 | 0.22 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr5_-_49417365 | 0.22 |
|
|
|
| chr20_+_28006043 | 0.22 |
|
|
|
| chr5_-_40503363 | 0.21 |
ENSDART00000074781
|
golph3
|
golgi phosphoprotein 3 |
| chr19_+_21783111 | 0.21 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr5_+_53851006 | 0.21 |
ENSDART00000157722
ENSDART00000160446 |
tmem203
|
transmembrane protein 203 |
| chr6_-_27118157 | 0.21 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr23_-_24556810 | 0.21 |
ENSDART00000109248
|
spen
|
spen family transcriptional repressor |
| chr15_-_3023693 | 0.21 |
|
|
|
| chr21_-_3548863 | 0.21 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr11_-_10867153 | 0.21 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr11_-_2437396 | 0.21 |
|
|
|
| chr14_-_1342450 | 0.20 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr3_-_39912816 | 0.20 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr9_+_42355513 | 0.20 |
ENSDART00000142888
|
lrrc3
|
leucine rich repeat containing 3 |
| chr14_+_15728850 | 0.20 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr22_+_1335355 | 0.20 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr20_-_29961621 | 0.20 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr1_+_50547385 | 0.19 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr11_-_6442490 | 0.19 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr16_-_42105733 | 0.19 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_-_52526843 | 0.19 |
ENSDART00000132941
|
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr20_-_20370323 | 0.19 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr13_+_26573404 | 0.19 |
ENSDART00000142483
|
fancl
|
Fanconi anemia, complementation group L |
| chr20_-_29961498 | 0.19 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr2_-_52792076 | 0.19 |
ENSDART00000097716
|
zgc:136336
|
zgc:136336 |
| chr18_-_43890836 | 0.18 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr16_+_34038263 | 0.18 |
ENSDART00000134946
|
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr10_+_33450122 | 0.18 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr16_-_16695773 | 0.18 |
ENSDART00000156294
|
CR848667.1
|
ENSDARG00000097712 |
| chr25_+_10450395 | 0.18 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr5_-_40503137 | 0.18 |
ENSDART00000051070
|
golph3
|
golgi phosphoprotein 3 |
| chr11_-_27378184 | 0.18 |
ENSDART00000157337
|
CR931782.1
|
ENSDARG00000097455 |
| chr18_+_8362131 | 0.18 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr7_-_19688306 | 0.18 |
ENSDART00000127669
|
prox1b
|
prospero homeobox 1b |
| chr16_+_23884575 | 0.17 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr7_+_16977507 | 0.17 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr13_+_32609784 | 0.17 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr24_-_14446593 | 0.17 |
|
|
|
| chr1_+_35253862 | 0.17 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr8_-_25015215 | 0.17 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr15_-_3023850 | 0.17 |
|
|
|
| chr10_-_44170848 | 0.17 |
ENSDART00000135240
|
acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr2_+_3115541 | 0.17 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr3_-_33296077 | 0.17 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr20_+_29306863 | 0.17 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_+_16038358 | 0.17 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr10_+_15297106 | 0.16 |
ENSDART00000139047
|
vldlr
|
very low density lipoprotein receptor |
| chr8_-_11950254 | 0.16 |
ENSDART00000005140
|
med27
|
mediator complex subunit 27 |
| chr20_-_23527234 | 0.16 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr20_-_29961589 | 0.16 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr5_-_3604175 | 0.16 |
ENSDART00000058346
|
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr14_+_30455165 | 0.16 |
ENSDART00000144817
|
cfl1
|
cofilin 1 |
| chr24_-_32864493 | 0.16 |
|
|
|
| chr17_-_37447917 | 0.16 |
ENSDART00000075975
|
crip1
|
cysteine-rich protein 1 |
| chr5_-_25133456 | 0.16 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr8_+_29733109 | 0.16 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| KN150037v1_+_2275 | 0.16 |
|
|
|
| chr22_-_26254217 | 0.16 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr13_+_32609849 | 0.15 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr3_+_18621758 | 0.15 |
ENSDART00000156747
|
AL772362.1
|
ENSDARG00000097552 |
| chr8_+_50964745 | 0.15 |
ENSDART00000013870
|
ENSDARG00000007359
|
ENSDARG00000007359 |
| chr25_-_95670 | 0.15 |
ENSDART00000163187
|
CABZ01083501.1
|
ENSDARG00000104429 |
| chr22_-_26254136 | 0.15 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr6_+_7257043 | 0.15 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr2_+_6341404 | 0.15 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_-_8585312 | 0.15 |
|
|
|
| chr9_-_32295031 | 0.15 |
ENSDART00000132448
|
BX530407.4
|
ENSDARG00000093950 |
| chr19_+_5562075 | 0.15 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr14_+_30073402 | 0.15 |
ENSDART00000136009
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr10_-_28230992 | 0.15 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr20_-_28898117 | 0.15 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr25_-_34631252 | 0.14 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr2_-_15656155 | 0.14 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr17_-_117828 | 0.14 |
|
|
|
| chr22_+_2074020 | 0.14 |
ENSDART00000106540
|
znf1161
|
zinc finger protein 1161 |
| chr14_-_36057494 | 0.14 |
ENSDART00000052562
|
spata4
|
spermatogenesis associated 4 |
| KN150456v1_-_19515 | 0.14 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr9_+_33310981 | 0.14 |
|
|
|
| chr16_+_17481157 | 0.14 |
ENSDART00000173448
|
fam131bb
|
family with sequence similarity 131, member Bb |
| chr3_+_18406137 | 0.14 |
ENSDART00000158791
|
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr16_-_25653129 | 0.14 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr3_+_27667194 | 0.14 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr18_+_14651021 | 0.14 |
ENSDART00000166679
|
cirh1a
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr25_-_24440292 | 0.14 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr14_+_8634323 | 0.14 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr11_+_31346456 | 0.14 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr15_-_4537178 | 0.14 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr12_-_33256671 | 0.13 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_-_33256934 | 0.13 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr4_+_75469488 | 0.13 |
ENSDART00000144892
|
znf1009
|
zinc finger protein 1009 |
| chr7_-_73590802 | 0.13 |
ENSDART00000167855
|
FP236812.8
|
Histone H2B 1/2 |
| chr16_-_20506304 | 0.13 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr25_+_25670946 | 0.13 |
ENSDART00000103638
|
idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr14_+_47326080 | 0.13 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_+_35163845 | 0.13 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr18_-_38787413 | 0.13 |
|
|
|
| chr6_+_6640324 | 0.13 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr2_+_1825714 | 0.13 |
ENSDART00000148624
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr9_+_18821640 | 0.13 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr22_-_19986461 | 0.13 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr22_-_19986607 | 0.13 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr8_+_48954402 | 0.13 |
|
|
|
| chr25_-_12236333 | 0.12 |
ENSDART00000174863
ENSDART00000179042 |
BX323452.1
|
ENSDARG00000107774 |
| chr14_-_33605295 | 0.12 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr14_-_26093969 | 0.12 |
ENSDART00000037999
|
b4galt7
|
xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.3 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.3 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.2 | GO:1903895 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.2 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 0.1 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 1.0 | GO:0098727 | maintenance of cell number(GO:0098727) |
| 0.0 | 0.1 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.7 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.9 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.1 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |