Project
DANIO-CODE
Navigation
Downloads
Results for hoxa5a
Z-value: 1.55
Transcription factors associated with hoxa5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa5a
|
ENSDARG00000102501 | homeobox A5a |
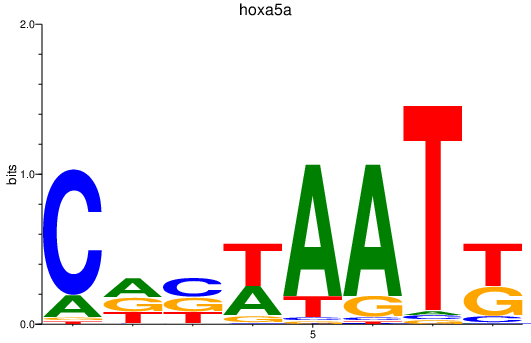
Activity profile of hoxa5a motif
Sorted Z-values of hoxa5a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa5a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_32747592 | 8.46 |
|
|
|
| chr7_+_17695289 | 8.40 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr12_-_13692190 | 4.85 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr24_-_14446593 | 4.80 |
|
|
|
| chr7_-_55346967 | 4.62 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr19_-_20819101 | 4.62 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr5_+_26888744 | 4.53 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr21_-_2296253 | 4.43 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr10_-_43070342 | 4.35 |
ENSDART00000099270
|
CU326366.2
|
ENSDARG00000068593 |
| chr11_-_44539778 | 4.03 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr22_-_38321005 | 3.91 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr2_-_44924505 | 3.16 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr10_-_1933874 | 3.04 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr1_-_43299713 | 2.93 |
ENSDART00000138447
|
zgc:66472
|
zgc:66472 |
| chr10_-_2915710 | 2.93 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr11_-_15886860 | 2.86 |
ENSDART00000170731
|
zgc:173544
|
zgc:173544 |
| chr21_-_32747544 | 2.80 |
|
|
|
| chr2_+_56534374 | 2.65 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr3_+_16692138 | 2.53 |
ENSDART00000023985
|
zgc:153952
|
zgc:153952 |
| chr10_+_44853207 | 2.48 |
ENSDART00000169466
|
scarb1
|
scavenger receptor class B, member 1 |
| chr21_+_43183522 | 2.42 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr16_-_39245082 | 2.34 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr12_-_10529153 | 2.16 |
ENSDART00000047769
|
myof
|
myoferlin |
| chr17_-_1524751 | 2.15 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr1_+_18909507 | 2.14 |
ENSDART00000088933
ENSDART00000141579 |
fbxo10
|
F-box protein 10 |
| chr17_-_38940072 | 2.07 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr23_+_32102030 | 2.01 |
ENSDART00000145501
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr1_+_18909404 | 1.98 |
ENSDART00000088933
ENSDART00000141579 |
fbxo10
|
F-box protein 10 |
| chr2_+_32863386 | 1.93 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr18_+_39079744 | 1.93 |
ENSDART00000098729
|
zgc:171509
|
zgc:171509 |
| chr12_-_10528974 | 1.92 |
ENSDART00000047769
|
myof
|
myoferlin |
| chr12_+_20222000 | 1.89 |
ENSDART00000150020
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr8_-_38284748 | 1.88 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr9_-_10174409 | 1.82 |
ENSDART00000004745
|
hnmt
|
histamine N-methyltransferase |
| chr21_-_2350090 | 1.80 |
ENSDART00000168946
|
si:ch211-241b2.4
|
si:ch211-241b2.4 |
| chr3_+_16692090 | 1.79 |
ENSDART00000023985
|
zgc:153952
|
zgc:153952 |
| chr17_+_5680342 | 1.77 |
ENSDART00000156089
|
CU929322.1
|
ENSDARG00000096839 |
| chr10_+_16914003 | 1.75 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr3_-_20944854 | 1.74 |
ENSDART00000109790
|
nlk1
|
nemo-like kinase, type 1 |
| chr10_-_109097 | 1.72 |
ENSDART00000127228
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr5_+_56988663 | 1.63 |
ENSDART00000074268
|
zgc:153929
|
zgc:153929 |
| chr4_-_5766814 | 1.61 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr8_-_39788989 | 1.59 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr7_+_26730804 | 1.58 |
|
|
|
| chr13_-_11873326 | 1.57 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr4_-_26106485 | 1.56 |
ENSDART00000100611
|
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr23_-_6831711 | 1.55 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr2_-_37821146 | 1.55 |
ENSDART00000154124
|
nfatc4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr4_-_17654047 | 1.52 |
|
|
|
| chr17_-_43041254 | 1.41 |
ENSDART00000124663
|
npc2
|
Niemann-Pick disease, type C2 |
| chr2_-_48444730 | 1.36 |
ENSDART00000057957
|
itm2cb
|
integral membrane protein 2Cb |
| chr22_-_26112676 | 1.32 |
|
|
|
| chr14_-_26199966 | 1.32 |
ENSDART00000054175
ENSDART00000145625 |
smad5
|
SMAD family member 5 |
| chr17_-_10581958 | 1.32 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr16_-_24916921 | 1.31 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr11_-_33605979 | 1.29 |
ENSDART00000171439
|
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr22_+_1445227 | 1.28 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr15_-_23756976 | 1.27 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr13_-_25620394 | 1.27 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr15_-_47351885 | 1.27 |
|
|
|
| chr11_-_6870810 | 1.26 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr7_+_73595871 | 1.25 |
ENSDART00000169756
|
HIST1H2BA (1 of many)
|
Histone H2B 1/2 |
| chr11_-_30388949 | 1.24 |
ENSDART00000063775
|
ENSDARG00000043442
|
ENSDARG00000043442 |
| chr14_-_26127173 | 1.20 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr1_-_52833426 | 1.19 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr19_+_7254917 | 1.18 |
ENSDART00000123934
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr6_+_6505309 | 1.17 |
ENSDART00000065568
|
zgc:113227
|
zgc:113227 |
| chr23_-_27647386 | 1.15 |
|
|
|
| chr12_+_22507388 | 1.15 |
ENSDART00000168935
|
mfsd8
|
major facilitator superfamily domain containing 8 |
| chr21_-_32747483 | 1.15 |
|
|
|
| chr6_+_23218068 | 1.14 |
ENSDART00000164960
|
stx8
|
syntaxin 8 |
| chr8_-_31706783 | 1.14 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr17_-_9806413 | 1.13 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr9_-_44797688 | 1.10 |
|
|
|
| chr10_-_10755962 | 1.09 |
ENSDART00000137797
|
CT027791.1
|
ENSDARG00000093881 |
| chr16_+_41004372 | 1.08 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr2_+_32842978 | 1.07 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr10_-_43070514 | 1.02 |
|
|
|
| chr13_-_25620444 | 1.01 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr17_+_16102236 | 0.99 |
ENSDART00000156989
|
CT954240.1
|
ENSDARG00000097742 |
| chr11_-_25019165 | 0.99 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr11_+_30389309 | 0.94 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr21_-_1618494 | 0.94 |
ENSDART00000124904
|
zgc:152948
|
zgc:152948 |
| chr12_-_7201660 | 0.93 |
ENSDART00000152450
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr9_-_12298632 | 0.93 |
ENSDART00000032344
|
nup35
|
nucleoporin 35 |
| chr7_-_40359613 | 0.92 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr1_-_43300015 | 0.92 |
ENSDART00000138447
|
zgc:66472
|
zgc:66472 |
| chr16_+_41004516 | 0.92 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr18_+_22286036 | 0.91 |
|
|
|
| chr24_-_14447519 | 0.90 |
|
|
|
| chr3_+_32234155 | 0.89 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr10_+_43070478 | 0.88 |
|
|
|
| chr17_-_27217309 | 0.85 |
ENSDART00000130080
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr11_-_44539726 | 0.83 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr11_+_3939876 | 0.81 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr22_+_37696341 | 0.81 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr21_-_2322963 | 0.81 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr2_-_37555425 | 0.81 |
ENSDART00000143496
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr8_+_45339021 | 0.81 |
|
|
|
| chr6_-_23966578 | 0.79 |
ENSDART00000164159
|
dr1
|
down-regulator of transcription 1 |
| chr21_-_35291005 | 0.77 |
ENSDART00000134780
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr11_+_30389137 | 0.76 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr11_-_7146819 | 0.75 |
ENSDART00000172879
|
smim7
|
small integral membrane protein 7 |
| chr15_-_44581550 | 0.74 |
|
|
|
| chr2_+_10209233 | 0.74 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr1_+_45147835 | 0.73 |
ENSDART00000179047
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr19_-_11001651 | 0.72 |
ENSDART00000165630
|
ecm1a
|
extracellular matrix protein 1a |
| chr25_+_33486711 | 0.71 |
ENSDART00000121498
|
ice2
|
interactor of little elongator complex ELL subunit 2 |
| chr2_+_8981028 | 0.69 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr6_+_23966336 | 0.68 |
|
|
|
| chr2_-_22913885 | 0.67 |
|
|
|
| chr16_-_39245193 | 0.66 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr7_+_71573058 | 0.64 |
ENSDART00000166865
|
myl12.1
|
myosin, light chain 12, genome duplicate 1 |
| chr13_-_16901290 | 0.64 |
|
|
|
| chr3_-_26927151 | 0.63 |
ENSDART00000153542
|
CR954963.1
|
ENSDARG00000097153 |
| chr3_+_27476229 | 0.56 |
ENSDART00000024453
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr2_-_31728072 | 0.53 |
ENSDART00000113498
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr15_+_17322173 | 0.52 |
ENSDART00000169608
|
dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr12_-_10529236 | 0.52 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr24_+_33916364 | 0.49 |
ENSDART00000136040
|
atg9b
|
autophagy related 9B |
| chr9_-_1046486 | 0.46 |
ENSDART00000161870
|
ino80da
|
INO80 complex subunit Da |
| chr11_-_37816180 | 0.46 |
ENSDART00000086516
|
klhdc8a
|
kelch domain containing 8A |
| chr9_-_746424 | 0.46 |
ENSDART00000082300
|
usp37
|
ubiquitin specific peptidase 37 |
| chr2_+_32843040 | 0.45 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr1_+_45147775 | 0.43 |
ENSDART00000179047
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr1_+_43347056 | 0.43 |
|
|
|
| chr19_-_4934737 | 0.42 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr4_-_7644374 | 0.41 |
ENSDART00000157751
|
BX649411.1
|
ENSDARG00000101254 |
| chr15_-_15294188 | 0.40 |
ENSDART00000155400
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr19_+_4939815 | 0.39 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr12_-_31609517 | 0.38 |
ENSDART00000080173
|
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr1_+_5492078 | 0.34 |
ENSDART00000140936
|
retreg2
|
reticulophagy regulator family member 2 |
| chr15_+_42329433 | 0.34 |
ENSDART00000130404
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr25_+_7172658 | 0.34 |
ENSDART00000111151
|
peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr14_-_1186858 | 0.33 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr8_-_38284660 | 0.31 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr2_+_24152953 | 0.29 |
ENSDART00000131030
|
gorasp1a
|
golgi reassembly stacking protein 1a |
| chr13_-_17598311 | 0.29 |
ENSDART00000013011
|
vdac2
|
voltage-dependent anion channel 2 |
| chr15_+_23588063 | 0.28 |
ENSDART00000152517
|
si:dkey-182i3.9
|
si:dkey-182i3.9 |
| chr21_+_17509116 | 0.28 |
|
|
|
| chr4_-_75660728 | 0.28 |
ENSDART00000154142
ENSDART00000155070 |
si:dkey-172k15.3
|
si:dkey-172k15.3 |
| chr14_+_10675644 | 0.24 |
ENSDART00000091169
|
abcb7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr16_+_10666588 | 0.22 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr13_-_49294664 | 0.17 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr19_+_43524684 | 0.13 |
|
|
|
| chr8_-_43733891 | 0.13 |
ENSDART00000166581
|
ep400
|
E1A binding protein p400 |
| chr10_+_5025384 | 0.13 |
ENSDART00000031165
|
enoph1
|
enolase-phosphatase 1 |
| chr16_-_2880109 | 0.11 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr8_-_23759198 | 0.11 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr11_+_10991763 | 0.11 |
ENSDART00000160488
|
itgb6
|
integrin, beta 6 |
| chr3_+_27475771 | 0.08 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr14_-_1186789 | 0.06 |
|
|
|
| chr16_-_21863723 | 0.00 |
ENSDART00000161640
|
gnl1
|
guanine nucleotide binding protein-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.0 | 4.8 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.9 | 4.6 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.8 | 3.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 1.8 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 | 4.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 1.4 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.3 | 1.3 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.2 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.6 | GO:0097065 | anterior head development(GO:0097065) |
| 0.2 | 2.0 | GO:0034381 | cholesterol efflux(GO:0033344) plasma lipoprotein particle clearance(GO:0034381) |
| 0.2 | 0.7 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 4.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 4.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.5 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.9 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 2.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.3 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 2.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.4 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.3 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 1.2 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.8 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 1.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.8 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.7 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 1.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.7 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.3 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.6 | GO:0031647 | regulation of protein stability(GO:0031647) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 5.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 0.8 | GO:0017054 | negative cofactor 2 complex(GO:0017054) |
| 0.1 | 0.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.2 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 2.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 3.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.5 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 4.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.7 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.6 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 4.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 2.0 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 2.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 0.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 3.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.3 | 1.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 5.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 2.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 4.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 4.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 1.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 2.0 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 3.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 2.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 2.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 4.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 7.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 2.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.1 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |