Project
DANIO-CODE
Navigation
Downloads
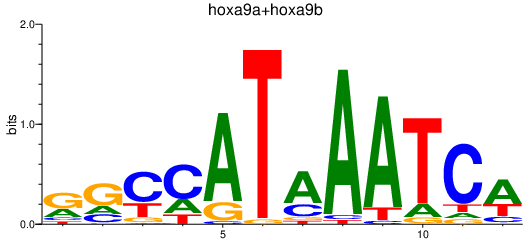
Results for hoxa9a+hoxa9b
Z-value: 1.41
Transcription factors associated with hoxa9a+hoxa9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa9b
|
ENSDARG00000056819 | homeobox A9b |
|
hoxa9a
|
ENSDARG00000105013 | homeobox A9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa9a | dr10_dc_chr19_+_20177128_20177172 | -0.91 | 8.1e-07 | Click! |
| hoxa9b | dr10_dc_chr16_+_21109486_21109575 | -0.90 | 1.5e-06 | Click! |
Activity profile of hoxa9a+hoxa9b motif
Sorted Z-values of hoxa9a+hoxa9b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa9a+hoxa9b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_32914487 | 5.78 |
ENSDART00000015547
|
cldng
|
claudin g |
| chr19_-_27966525 | 3.71 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr8_-_2557556 | 3.51 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| KN150456v1_-_19515 | 3.48 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr2_+_34984631 | 3.10 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr7_+_30355221 | 2.94 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr20_-_20922072 | 2.87 |
ENSDART00000142618
ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr17_+_19606608 | 2.83 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr17_-_4086835 | 2.80 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr5_-_23211957 | 2.73 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr12_-_22116694 | 2.66 |
ENSDART00000038310
ENSDART00000132731 |
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr14_+_34150130 | 2.59 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr2_+_58943872 | 2.56 |
ENSDART00000158860
ENSDART00000067736 |
stk11
|
serine/threonine kinase 11 |
| chr10_+_44853207 | 2.36 |
ENSDART00000169466
|
scarb1
|
scavenger receptor class B, member 1 |
| chr19_-_15324823 | 2.29 |
ENSDART00000169883
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr6_-_40451790 | 2.26 |
ENSDART00000103879
|
tatdn2
|
TatD DNase domain containing 2 |
| chr21_-_13593659 | 2.20 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr1_+_23866532 | 2.17 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr14_+_32578253 | 2.13 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr25_-_36512943 | 2.10 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| KN150623v1_+_258 | 2.10 |
|
|
|
| chr11_-_22663430 | 2.08 |
ENSDART00000154813
|
mdm4
|
MDM4, p53 regulator |
| chr7_+_26274687 | 2.04 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr8_-_38322610 | 2.01 |
ENSDART00000134283
ENSDART00000132077 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr25_-_2932779 | 1.98 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr6_-_25065823 | 1.91 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr3_-_39316317 | 1.90 |
|
|
|
| chr18_+_48188699 | 1.88 |
|
|
|
| chr24_-_32259029 | 1.86 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr14_-_32937536 | 1.85 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr21_+_15774697 | 1.84 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr21_+_15774822 | 1.79 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr3_+_44928323 | 1.78 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr7_-_37648061 | 1.77 |
ENSDART00000052368
|
heatr3
|
HEAT repeat containing 3 |
| chr23_+_29017954 | 1.76 |
ENSDART00000140291
|
CR677513.1
|
ENSDARG00000093890 |
| chr12_-_25058837 | 1.76 |
ENSDART00000135368
|
rhoq
|
ras homolog family member Q |
| chr22_+_21524515 | 1.74 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr7_+_1337856 | 1.72 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr7_-_19116999 | 1.71 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr23_-_3778250 | 1.70 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr21_+_19958025 | 1.70 |
|
|
|
| chr1_-_51567135 | 1.70 |
ENSDART00000143805
|
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr13_+_13814142 | 1.67 |
ENSDART00000142997
|
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr3_+_40113121 | 1.66 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr15_+_24629719 | 1.65 |
ENSDART00000134622
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr20_-_23355244 | 1.65 |
|
|
|
| chr21_-_3548863 | 1.63 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr5_-_66145078 | 1.62 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr6_-_42390995 | 1.61 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr3_-_40090673 | 1.60 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr15_+_24629858 | 1.56 |
ENSDART00000134622
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr16_-_43107682 | 1.51 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr11_-_22663375 | 1.50 |
ENSDART00000154813
|
mdm4
|
MDM4, p53 regulator |
| chr7_+_13742576 | 1.50 |
|
|
|
| KN150334v1_-_9585 | 1.48 |
ENSDART00000175935
|
CABZ01113810.1
|
ENSDARG00000107898 |
| chr20_+_54404987 | 1.47 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr25_-_24440001 | 1.44 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr9_-_2522639 | 1.44 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr5_+_19429620 | 1.43 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr22_+_25754857 | 1.42 |
ENSDART00000174421
|
AL929192.1
|
ENSDARG00000106684 |
| chr20_-_34851706 | 1.41 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr14_-_31375046 | 1.39 |
ENSDART00000173274
|
map7d3
|
MAP7 domain containing 3 |
| chr3_-_26060787 | 1.38 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr8_-_2557506 | 1.37 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr9_-_48516764 | 1.36 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr10_-_1933761 | 1.35 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr16_-_34240818 | 1.35 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr5_+_19429500 | 1.35 |
ENSDART00000168868
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr1_+_30925269 | 1.34 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr25_-_2932820 | 1.34 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr6_-_25065745 | 1.34 |
ENSDART00000165170
|
znf326
|
zinc finger protein 326 |
| chr17_+_15780112 | 1.33 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr21_-_20908880 | 1.32 |
ENSDART00000079701
|
rnf180
|
ring finger protein 180 |
| chr17_+_15780156 | 1.31 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr16_+_28335377 | 1.31 |
ENSDART00000059035
|
fam188a
|
family with sequence similarity 188, member A |
| chr9_-_30448622 | 1.30 |
ENSDART00000129926
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr3_+_13034056 | 1.28 |
|
|
|
| chr17_+_25313170 | 1.28 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr7_-_19989419 | 1.28 |
ENSDART00000127699
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr25_+_15901398 | 1.28 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr10_-_8088063 | 1.25 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr5_-_31796734 | 1.25 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr12_-_7551118 | 1.24 |
ENSDART00000152556
|
FAM13C
|
family with sequence similarity 13 member C |
| chr23_-_39253291 | 1.24 |
|
|
|
| chr1_+_57267909 | 1.24 |
ENSDART00000152640
|
BX469930.2
|
ENSDARG00000096615 |
| chr7_-_38363533 | 1.23 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr11_+_14285767 | 1.23 |
ENSDART00000171347
|
si:ch211-262i1.6
|
si:ch211-262i1.6 |
| chr3_+_58417512 | 1.22 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr7_+_13742622 | 1.22 |
|
|
|
| chr8_+_3373066 | 1.22 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr1_+_35253862 | 1.21 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr17_-_4086971 | 1.20 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr8_-_44247277 | 1.20 |
|
|
|
| chr17_+_11519212 | 1.20 |
|
|
|
| chr2_-_58805035 | 1.20 |
ENSDART00000159735
|
mau2
|
MAU2 sister chromatid cohesion factor |
| chr3_-_14545237 | 1.19 |
ENSDART00000133850
|
gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr24_-_6304289 | 1.18 |
ENSDART00000140212
|
zgc:174877
|
zgc:174877 |
| chr19_-_35286549 | 1.18 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr6_-_53145759 | 1.18 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr23_-_43916621 | 1.18 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr1_-_54505313 | 1.17 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr14_+_31265616 | 1.15 |
|
|
|
| chr12_+_29017351 | 1.12 |
|
|
|
| chr22_+_11114268 | 1.12 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
| chr4_+_16197 | 1.12 |
ENSDART00000166174
|
ENSDARG00000100660
|
ENSDARG00000100660 |
| chr5_-_40524177 | 1.12 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr15_+_24629778 | 1.11 |
ENSDART00000134622
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr16_-_17678748 | 1.11 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr16_-_34471672 | 1.10 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr24_+_26257058 | 1.10 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr3_+_9504996 | 1.09 |
ENSDART00000171467
|
si:dkey-29p9.3
|
si:dkey-29p9.3 |
| chr16_+_32060609 | 1.08 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr13_+_51485084 | 1.08 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr13_-_35782121 | 1.05 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr1_+_30925353 | 1.04 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr3_-_59690168 | 1.04 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr2_-_44924505 | 1.03 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr20_-_52002087 | 1.03 |
ENSDART00000041476
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr6_+_18894174 | 1.02 |
ENSDART00000165806
|
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr5_-_35997345 | 1.01 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr8_+_15231620 | 1.01 |
ENSDART00000020386
|
gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr15_-_37037547 | 1.01 |
|
|
|
| chr14_+_25950198 | 0.97 |
ENSDART00000113804
ENSDART00000159054 |
CCDC69
|
coiled-coil domain containing 69 |
| chr18_-_35432838 | 0.97 |
ENSDART00000141703
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr3_+_58417635 | 0.96 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr8_+_25072241 | 0.96 |
ENSDART00000143922
|
atxn7l2b
|
ataxin 7-like 2b |
| chr22_-_11024649 | 0.95 |
ENSDART00000105823
ENSDART00000159995 |
insrb
|
insulin receptor b |
| chr20_+_29685379 | 0.95 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr18_-_35432655 | 0.95 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr25_+_3169073 | 0.93 |
|
|
|
| chr22_+_11114168 | 0.93 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
| chr10_+_33758581 | 0.93 |
ENSDART00000141650
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr12_+_4651222 | 0.93 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr15_+_29241472 | 0.92 |
|
|
|
| chr11_-_13283709 | 0.92 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr3_-_40112966 | 0.92 |
ENSDART00000154562
|
top3a
|
topoisomerase (DNA) III alpha |
| chr17_-_2419079 | 0.91 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr18_+_41559512 | 0.91 |
ENSDART00000059135
|
bcl7bb
|
B-cell CLL/lymphoma 7B, b |
| chr13_-_22831005 | 0.90 |
ENSDART00000143112
|
tspan15
|
tetraspanin 15 |
| chr23_+_3778066 | 0.90 |
ENSDART00000141880
|
smim29
|
small integral membrane protein 29 |
| chr24_-_20946041 | 0.90 |
ENSDART00000140786
|
qtrtd1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr10_-_1933874 | 0.90 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr6_-_11576632 | 0.89 |
ENSDART00000151717
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr16_+_29574449 | 0.87 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr1_-_529071 | 0.87 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr3_-_40626599 | 0.87 |
ENSDART00000004923
|
smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr23_+_43916520 | 0.87 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr19_-_34529880 | 0.87 |
ENSDART00000158677
ENSDART00000112004 |
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr8_-_38322559 | 0.87 |
ENSDART00000134283
ENSDART00000132077 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr13_-_24778226 | 0.86 |
|
|
|
| KN149698v1_-_88818 | 0.86 |
|
|
|
| chr6_-_14841372 | 0.86 |
ENSDART00000167436
|
CU929521.1
|
ENSDARG00000097482 |
| chr13_+_13814262 | 0.86 |
ENSDART00000142997
|
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr1_+_11490451 | 0.86 |
ENSDART00000142081
|
stra6l
|
STRA6-like |
| chr13_-_24778352 | 0.85 |
|
|
|
| chr5_-_28914505 | 0.85 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr10_-_25448712 | 0.85 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr20_-_54812675 | 0.84 |
ENSDART00000059872
|
ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr17_-_25612341 | 0.84 |
ENSDART00000126201
ENSDART00000105503 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr8_+_3372903 | 0.84 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr13_-_22831037 | 0.83 |
ENSDART00000057641
|
tspan15
|
tetraspanin 15 |
| chr2_-_32755182 | 0.83 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr2_+_21527785 | 0.83 |
ENSDART00000136498
|
si:dkey-29d8.3
|
si:dkey-29d8.3 |
| chr19_+_3115685 | 0.83 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr7_+_38257950 | 0.82 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr12_+_13050549 | 0.82 |
|
|
|
| chr11_-_44107913 | 0.82 |
|
|
|
| chr16_+_29442108 | 0.81 |
ENSDART00000050535
|
rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr3_+_52290252 | 0.81 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr15_-_14653696 | 0.81 |
ENSDART00000172195
|
adck4
|
aarF domain containing kinase 4 |
| chr11_+_5522377 | 0.81 |
ENSDART00000013203
|
cse1l
|
CSE1 chromosome segregation 1-like (yeast) |
| chr10_-_33307845 | 0.81 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr25_+_16784585 | 0.80 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr5_+_63596241 | 0.80 |
ENSDART00000111282
|
qsox2
|
quiescin Q6 sulfhydryl oxidase 2 |
| KN150604v1_-_7233 | 0.80 |
|
|
|
| chr6_-_53145582 | 0.80 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr19_-_15324909 | 0.79 |
ENSDART00000169883
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr2_+_35613093 | 0.79 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr2_+_22753634 | 0.79 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr5_+_55763413 | 0.79 |
|
|
|
| chr11_-_40192951 | 0.78 |
ENSDART00000102750
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr3_+_52290504 | 0.78 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr11_-_16261061 | 0.78 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr5_+_61279071 | 0.78 |
ENSDART00000168808
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr22_-_9796735 | 0.78 |
ENSDART00000144395
|
BX539307.3
|
ENSDARG00000093730 |
| chr20_+_2625716 | 0.77 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr25_+_2252667 | 0.77 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr22_+_21524430 | 0.77 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr16_-_9939549 | 0.77 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr9_-_50106512 | 0.77 |
|
|
|
| chr17_-_25612397 | 0.76 |
ENSDART00000126201
ENSDART00000105503 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr10_-_33307992 | 0.76 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr15_-_1519615 | 0.76 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr22_+_1643493 | 0.76 |
ENSDART00000167767
|
BX649416.1
|
ENSDARG00000098230 |
| chr23_-_3778530 | 0.76 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr16_-_34471475 | 0.76 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr11_+_23796452 | 0.76 |
ENSDART00000128309
|
zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr3_+_24510974 | 0.75 |
ENSDART00000148414
ENSDART00000055590 |
zgc:113411
|
zgc:113411 |
| chr19_-_31455278 | 0.75 |
ENSDART00000133101
ENSDART00000136213 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr16_-_29517367 | 0.74 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.9 | 2.7 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.7 | 2.2 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.6 | 2.6 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 3.1 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.5 | 2.2 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.5 | 1.6 | GO:0051055 | medium-chain fatty acid transport(GO:0001579) temperature homeostasis(GO:0001659) negative regulation of lipid biosynthetic process(GO:0051055) |
| 0.5 | 2.1 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.5 | 1.5 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.5 | 2.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 3.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 2.9 | GO:1901911 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.4 | 2.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 1.2 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.4 | 2.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.4 | 2.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 2.9 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 1.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.3 | 4.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.3 | 0.9 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.3 | 1.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 1.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 0.8 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 2.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 2.4 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.3 | 1.9 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.3 | 1.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.7 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.2 | 5.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 2.0 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 1.3 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.2 | 0.6 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 1.8 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.2 | 1.6 | GO:0031204 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.7 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.8 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 0.4 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.8 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.2 | GO:1901797 | negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 | 0.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 3.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 4.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 2.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.8 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 2.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 2.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.8 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 1.0 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 2.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 1.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 3.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 2.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 2.9 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.0 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.0 | GO:1902622 | engulfment of apoptotic cell(GO:0043652) regulation of neutrophil migration(GO:1902622) |
| 0.1 | 0.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0072019 | homeostasis of number of cells within a tissue(GO:0048873) proximal convoluted tubule development(GO:0072019) |
| 0.0 | 0.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 1.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.8 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.7 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 1.1 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 1.8 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.6 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.5 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.3 | GO:0006213 | pyrimidine nucleoside metabolic process(GO:0006213) |
| 0.0 | 0.2 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.8 | GO:0032259 | methylation(GO:0032259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.9 | 2.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.6 | 2.6 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.6 | 2.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 1.6 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 2.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 0.6 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 0.6 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 1.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.6 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 2.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 3.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0017054 | negative cofactor 2 complex(GO:0017054) |
| 0.0 | 0.9 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.7 | 2.1 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.5 | 2.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.5 | 1.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 2.9 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.4 | 1.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 2.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 2.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 1.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 0.7 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.2 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.9 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 0.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 2.2 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 1.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.6 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 1.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 3.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 3.1 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 2.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 1.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 2.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.0 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.7 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 3.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 6.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.1 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.6 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 2.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 2.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 5.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 3.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 3.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.7 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 6.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 4.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.2 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 0.0 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.6 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 4.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 5.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 5.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 3.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.3 | 1.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 2.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 2.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 1.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 2.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.4 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |