Project
DANIO-CODE
Navigation
Downloads
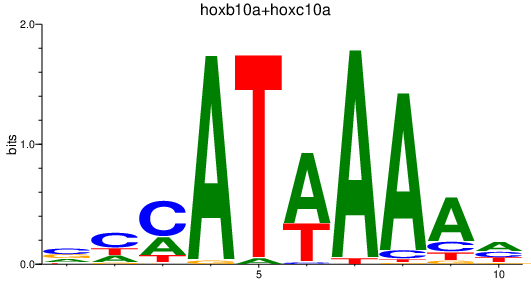
Results for hoxb10a+hoxc10a
Z-value: 0.47
Transcription factors associated with hoxb10a+hoxc10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb10a
|
ENSDARG00000011579 | homeobox B10a |
|
hoxc10a
|
ENSDARG00000070348 | homeobox C10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc10a | dr10_dc_chr23_+_35984969_35984991 | 0.77 | 5.5e-04 | Click! |
| hoxb10a | dr10_dc_chr3_+_23538277_23538283 | 0.68 | 3.5e-03 | Click! |
Activity profile of hoxb10a+hoxc10a motif
Sorted Z-values of hoxb10a+hoxc10a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb10a+hoxc10a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45950530 | 1.89 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr23_+_10211543 | 1.35 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr1_-_37475807 | 1.19 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr14_-_24463906 | 1.06 |
ENSDART00000126199
|
slit3
|
slit homolog 3 (Drosophila) |
| chr25_-_13518274 | 0.97 |
ENSDART00000165510
|
fa2h
|
fatty acid 2-hydroxylase |
| chr23_+_27141681 | 0.96 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr8_+_2428689 | 0.96 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr23_+_39713307 | 0.90 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr19_+_22478256 | 0.84 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr12_+_13080209 | 0.84 |
ENSDART00000127870
|
cmn
|
calymmin |
| chr2_-_35584440 | 0.83 |
ENSDART00000125298
|
tnw
|
tenascin W |
| chr25_-_30845998 | 0.83 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr17_-_20267580 | 0.83 |
ENSDART00000078703
|
add3b
|
adducin 3 (gamma) b |
| chr19_+_17926983 | 0.79 |
ENSDART00000141397
|
ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr25_+_19992389 | 0.76 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr16_-_33105847 | 0.75 |
|
|
|
| chr23_+_25428372 | 0.75 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr3_-_31672763 | 0.73 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr6_+_52791515 | 0.72 |
ENSDART00000065682
|
matn4
|
matrilin 4 |
| chr23_+_22977031 | 0.71 |
|
|
|
| chr20_+_26639029 | 0.71 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr23_+_7445760 | 0.67 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr5_+_36010448 | 0.66 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr25_+_30701751 | 0.66 |
|
|
|
| chr19_-_39048324 | 0.65 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr6_+_35378839 | 0.64 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr19_+_42899678 | 0.62 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr14_+_45895103 | 0.60 |
|
|
|
| chr6_-_28231995 | 0.59 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr18_+_22617072 | 0.59 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr23_-_27645138 | 0.57 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr16_+_28819826 | 0.57 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr25_+_34069931 | 0.56 |
|
|
|
| chr23_+_24711233 | 0.56 |
|
|
|
| chr8_+_18552850 | 0.56 |
ENSDART00000177476
|
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr2_+_53253623 | 0.55 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr6_-_60152693 | 0.54 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr6_-_60152594 | 0.53 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr2_+_20748431 | 0.53 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr22_+_4791383 | 0.52 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr9_-_32942783 | 0.52 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr10_-_30016761 | 0.51 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr1_-_50215233 | 0.50 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr19_-_17926919 | 0.49 |
ENSDART00000167592
ENSDART00000162383 |
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr21_-_20291707 | 0.49 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_-_20441931 | 0.48 |
|
|
|
| chr12_+_27026112 | 0.47 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr19_-_15951003 | 0.47 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr7_+_31608878 | 0.44 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_+_22134507 | 0.43 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr23_-_18203680 | 0.42 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr23_-_29138952 | 0.41 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr20_+_9776103 | 0.39 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr19_-_10962536 | 0.38 |
ENSDART00000160438
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr2_+_1881022 | 0.38 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr22_-_22696091 | 0.34 |
|
|
|
| chr10_+_22876094 | 0.34 |
|
|
|
| chr10_+_2559774 | 0.33 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr20_+_32646617 | 0.33 |
|
|
|
| chr5_+_26312951 | 0.33 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr13_+_12257610 | 0.33 |
ENSDART00000086525
|
atp10d
|
ATPase, class V, type 10D |
| chr19_-_39048402 | 0.32 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr21_+_10663517 | 0.32 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr19_-_46058895 | 0.30 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr19_-_46058954 | 0.30 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr17_-_2513630 | 0.30 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr6_+_60152913 | 0.30 |
|
|
|
| chr19_+_20189224 | 0.29 |
ENSDART00000163611
|
hoxa4a
|
homeobox A4a |
| chr24_-_8592157 | 0.29 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr23_-_35997065 | 0.28 |
ENSDART00000153910
|
BX005254.4
|
ENSDARG00000097150 |
| chr25_-_13579869 | 0.27 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr6_-_47386720 | 0.27 |
ENSDART00000058736
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr2_-_6127811 | 0.26 |
ENSDART00000013079
|
scp2a
|
sterol carrier protein 2a |
| chr13_+_36459706 | 0.25 |
ENSDART00000138940
|
gmfb
|
glia maturation factor, beta |
| chr6_+_30716955 | 0.24 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr10_-_22875997 | 0.24 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr10_+_9762039 | 0.24 |
|
|
|
| chr10_-_38206060 | 0.24 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr11_+_35896325 | 0.24 |
ENSDART00000163330
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr23_+_11734349 | 0.23 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr3_+_54506912 | 0.23 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr25_+_5567388 | 0.23 |
|
|
|
| chr12_-_20251183 | 0.23 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr1_+_50763649 | 0.22 |
ENSDART00000074294
|
actr2a
|
ARP2 actin related protein 2a homolog |
| chr11_-_18166056 | 0.22 |
ENSDART00000155752
|
sfmbt1
|
Scm-like with four mbt domains 1 |
| chr3_+_26015378 | 0.22 |
|
|
|
| chr11_+_25495396 | 0.22 |
|
|
|
| chr23_+_22858531 | 0.22 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr14_+_20862466 | 0.20 |
ENSDART00000059796
|
ENSDARG00000019213
|
ENSDARG00000019213 |
| chr24_-_8592102 | 0.19 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr16_+_13993746 | 0.19 |
ENSDART00000101304
|
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr9_+_30909382 | 0.19 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr2_+_33907298 | 0.19 |
|
|
|
| chr20_+_52740555 | 0.19 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr11_+_7570097 | 0.18 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr9_+_30909291 | 0.18 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr3_+_24003840 | 0.18 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr4_+_28452948 | 0.18 |
ENSDART00000150879
|
znf1067
|
zinc finger protein 1067 |
| chr11_+_18020191 | 0.17 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr18_-_15361133 | 0.17 |
ENSDART00000019818
|
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr11_+_7570027 | 0.17 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr5_+_45823481 | 0.17 |
|
|
|
| chr18_+_41242498 | 0.16 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr5_+_14701877 | 0.16 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr21_-_25719352 | 0.16 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr2_+_16928379 | 0.16 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr14_-_2342593 | 0.15 |
|
|
|
| chr14_+_45895037 | 0.15 |
|
|
|
| chr21_+_17509116 | 0.15 |
|
|
|
| chr5_+_22632707 | 0.14 |
ENSDART00000003428
|
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr23_+_22858773 | 0.13 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr9_+_41223155 | 0.13 |
ENSDART00000014660
|
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr11_+_2630587 | 0.13 |
ENSDART00000082512
|
tmem167b
|
transmembrane protein 167B |
| chr4_-_29488075 | 0.13 |
ENSDART00000167636
|
CR847895.4
|
ENSDARG00000101492 |
| chr15_-_1880375 | 0.12 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_23741791 | 0.11 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr14_-_47226179 | 0.10 |
ENSDART00000056734
ENSDART00000143158 |
setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr18_+_20836320 | 0.10 |
ENSDART00000090100
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr1_+_33510660 | 0.10 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr2_-_26160882 | 0.09 |
ENSDART00000133163
|
CR392026.2
|
ENSDARG00000094803 |
| chr9_+_1968455 | 0.09 |
|
|
|
| chr4_-_16461748 | 0.08 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr23_-_10906094 | 0.08 |
|
|
|
| chr4_-_16887296 | 0.07 |
ENSDART00000044005
ENSDART00000042874 ENSDART00000016690 |
tmpoa
|
thymopoietin a |
| chr8_+_8822226 | 0.07 |
ENSDART00000081999
|
otud5a
|
OTU deubiquitinase 5a |
| chr23_+_22858361 | 0.07 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr11_-_27970634 | 0.06 |
|
|
|
| chr6_+_12849629 | 0.06 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr16_+_25749018 | 0.06 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr8_+_23172960 | 0.06 |
ENSDART00000028946
|
tpd52l2a
|
tumor protein D52-like 2a |
| chr10_+_39142185 | 0.06 |
|
|
|
| chr23_+_36607999 | 0.06 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr25_+_19992328 | 0.06 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr10_+_18921391 | 0.06 |
ENSDART00000024127
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr12_+_31523317 | 0.05 |
ENSDART00000153129
|
dnmbp
|
dynamin binding protein |
| chr5_-_49724579 | 0.05 |
ENSDART00000157800
|
si:ch73-280o22.2
|
si:ch73-280o22.2 |
| chr1_-_51658383 | 0.05 |
ENSDART00000132638
|
ENSDARG00000059948
|
ENSDARG00000059948 |
| chr19_+_20189194 | 0.04 |
ENSDART00000163611
|
hoxa4a
|
homeobox A4a |
| chr17_-_48833163 | 0.04 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr21_+_15637884 | 0.04 |
ENSDART00000149371
|
idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr21_-_11877525 | 0.03 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr19_+_2659309 | 0.02 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr13_+_24453943 | 0.02 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr3_+_24004141 | 0.02 |
ENSDART00000155216
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr14_+_23419894 | 0.01 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_-_43896998 | 0.01 |
ENSDART00000156513
|
BX465227.1
|
ENSDARG00000097825 |
| chr1_-_17073539 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 1.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 1.0 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.5 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 0.7 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 1.0 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.6 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0043282 | pharyngeal muscle development(GO:0043282) |
| 0.1 | 1.9 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.2 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.5 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 1.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.7 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.5 | GO:0050935 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) iridophore differentiation(GO:0050935) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.0 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.2 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |