Project
DANIO-CODE
Navigation
Downloads
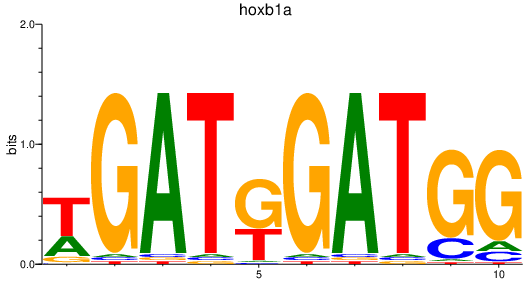
Results for hoxb1a
Z-value: 1.40
Transcription factors associated with hoxb1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb1a
|
ENSDARG00000008174 | homeobox B1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb1a | dr10_dc_chr3_+_23638277_23638323 | 0.86 | 1.7e-05 | Click! |
Activity profile of hoxb1a motif
Sorted Z-values of hoxb1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_23638277 | 6.92 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr6_+_29800606 | 4.58 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr18_+_40364675 | 4.29 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr5_-_35729429 | 4.20 |
|
|
|
| chr21_-_28883441 | 3.91 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr11_+_36933891 | 3.51 |
ENSDART00000170542
|
ENSDARG00000100406
|
ENSDARG00000100406 |
| chr1_-_37990863 | 3.31 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr23_+_36079164 | 3.27 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr7_+_42124857 | 3.27 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr14_-_36037883 | 3.15 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr18_+_24935499 | 3.04 |
|
|
|
| chr20_-_35938683 | 2.99 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr11_-_27455242 | 2.90 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr23_+_36079624 | 2.86 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr3_+_15656123 | 2.83 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr5_+_6054781 | 2.82 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr25_+_20118286 | 2.54 |
ENSDART00000104297
|
tnnt2d
|
troponin T2d, cardiac |
| chr7_+_16583234 | 2.52 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr11_-_27455348 | 2.37 |
ENSDART00000045942
|
phf2
|
PHD finger protein 2 |
| chr1_+_45503061 | 2.24 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr8_-_38168395 | 2.18 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_25378105 | 2.13 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr1_-_20218263 | 2.06 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr17_+_52736192 | 2.01 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr16_+_46145286 | 2.01 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr16_+_51317933 | 1.95 |
ENSDART00000157736
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr12_+_27444832 | 1.90 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr16_+_46435014 | 1.88 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr10_-_44509732 | 1.85 |
|
|
|
| chr10_+_44509370 | 1.80 |
|
|
|
| chr13_+_25722545 | 1.75 |
|
|
|
| chr14_+_34207218 | 1.72 |
ENSDART00000135608
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr1_-_40536800 | 1.72 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr24_+_34227202 | 1.68 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr22_+_35113233 | 1.58 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr16_+_41922320 | 1.58 |
|
|
|
| chr10_+_17494274 | 1.56 |
|
|
|
| chr4_+_26507297 | 1.50 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr4_-_8005840 | 1.44 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr21_+_16987306 | 1.44 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr8_+_5222065 | 1.43 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr12_-_25825072 | 1.38 |
|
|
|
| chr5_+_16669696 | 1.37 |
|
|
|
| chr5_+_35729588 | 1.35 |
|
|
|
| chr1_-_37990935 | 1.25 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr1_-_25377787 | 1.22 |
|
|
|
| chr23_-_19053587 | 1.21 |
|
|
|
| chr3_-_55399331 | 1.21 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr16_+_21121428 | 1.20 |
|
|
|
| chr1_+_23093114 | 1.16 |
|
|
|
| chr23_+_27830375 | 1.13 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr17_-_19606453 | 1.11 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr1_-_20132900 | 1.08 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr1_-_7744605 | 1.06 |
ENSDART00000033917
|
syngr3b
|
synaptogyrin 3b |
| chr22_-_16971180 | 1.03 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr2_+_56946090 | 1.01 |
|
|
|
| chr7_+_36627685 | 1.01 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr7_-_13128257 | 0.99 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr5_-_33156615 | 0.98 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr2_+_56946287 | 0.97 |
|
|
|
| chr22_-_16971035 | 0.97 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr21_-_28883243 | 0.93 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr17_+_23102259 | 0.88 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr13_+_26943081 | 0.87 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr7_-_33078758 | 0.84 |
ENSDART00000008785
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr18_+_41552295 | 0.78 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr17_-_50899135 | 0.77 |
|
|
|
| chr10_-_25629613 | 0.68 |
ENSDART00000131640
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr7_+_16583304 | 0.66 |
ENSDART00000113332
|
nav2a
|
neuron navigator 2a |
| chr22_-_16970979 | 0.66 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr1_+_23093475 | 0.65 |
|
|
|
| chr7_-_68229145 | 0.61 |
|
|
|
| chr16_-_1479139 | 0.61 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr7_+_33079046 | 0.59 |
|
|
|
| chr19_-_42759951 | 0.59 |
|
|
|
| chr14_+_43599635 | 0.57 |
ENSDART00000155539
|
CR786577.1
|
ENSDARG00000097875 |
| chr14_-_26078923 | 0.53 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr2_+_56946128 | 0.52 |
|
|
|
| chr4_-_8005933 | 0.52 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr7_+_29681510 | 0.51 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr2_-_16548451 | 0.51 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr21_+_32285006 | 0.50 |
|
|
|
| chr7_-_46510731 | 0.50 |
|
|
|
| chr17_-_28084988 | 0.46 |
ENSDART00000149654
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr16_-_31230568 | 0.45 |
ENSDART00000058829
|
scrt1b
|
scratch family zinc finger 1b |
| chr2_-_53797472 | 0.45 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr7_+_36627787 | 0.42 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr8_-_17692185 | 0.42 |
|
|
|
| chr1_-_37991246 | 0.35 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr23_+_22645604 | 0.32 |
ENSDART00000177273
|
CR388207.2
|
ENSDARG00000108862 |
| chr14_-_25645760 | 0.29 |
ENSDART00000141916
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr16_+_51696466 | 0.29 |
|
|
|
| chr12_+_27444762 | 0.28 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr25_+_7287952 | 0.27 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr2_-_53797503 | 0.27 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr7_-_68229018 | 0.25 |
|
|
|
| chr5_+_8460247 | 0.25 |
ENSDART00000091397
|
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr5_-_51631013 | 0.24 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr18_-_40763343 | 0.23 |
ENSDART00000130397
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_+_1881022 | 0.21 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr16_+_41922407 | 0.17 |
|
|
|
| chr17_+_23102283 | 0.12 |
ENSDART00000138069
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_686007 | 0.10 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr1_-_20133116 | 0.09 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr22_-_562269 | 0.07 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr11_-_23333850 | 0.06 |
|
|
|
| chr21_-_28883200 | 0.04 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr20_-_37673852 | 0.03 |
|
|
|
| chr10_+_37238790 | 0.03 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr17_+_51675406 | 0.02 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr19_+_13502773 | 0.01 |
ENSDART00000157631
|
si:ch211-204a13.2
|
si:ch211-204a13.2 |
| chr15_-_46439102 | 0.01 |
ENSDART00000125211
|
ENSDARG00000087950
|
ENSDARG00000087950 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 1.4 | 6.9 | GO:0021561 | facial nerve development(GO:0021561) |
| 1.0 | 3.0 | GO:0050864 | regulation of B cell activation(GO:0050864) |
| 0.3 | 4.9 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.3 | 1.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 1.7 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.1 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 2.8 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 0.6 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 1.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.5 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 4.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.2 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 2.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 2.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 4.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 2.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 3.4 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.0 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.0 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 2.1 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 2.7 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 5.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 6.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.1 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.5 | 4.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 5.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 2.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 4.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.8 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 23.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 4.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |