Project
DANIO-CODE
Navigation
Downloads
Results for hoxb2a
Z-value: 0.74
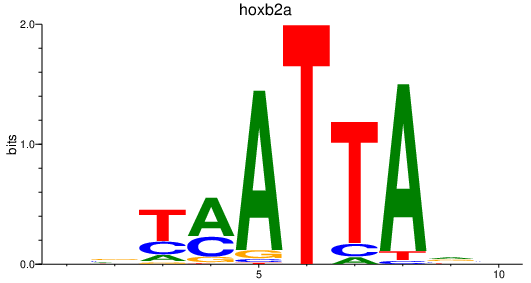
Transcription factors associated with hoxb2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb2a
|
ENSDARG00000000175 | homeobox B2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb2a | dr10_dc_chr3_+_23621843_23621906 | -0.27 | 3.2e-01 | Click! |
Activity profile of hoxb2a motif
Sorted Z-values of hoxb2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25740782 | 1.58 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr11_-_6442588 | 1.18 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr8_-_23759076 | 0.93 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr18_+_20571619 | 0.92 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr10_-_44713495 | 0.88 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr20_+_37023072 | 0.82 |
ENSDART00000155058
|
CR388421.1
|
ENSDARG00000096706 |
| chr23_+_28396415 | 0.76 |
ENSDART00000142179
|
birc5b
|
baculoviral IAP repeat containing 5b |
| KN150699v1_-_15078 | 0.74 |
ENSDART00000159861
|
ENSDARG00000098739
|
ENSDARG00000098739 |
| chr5_+_29193876 | 0.73 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr18_+_6417959 | 0.72 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr21_-_32027717 | 0.72 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr5_+_58936399 | 0.70 |
|
|
|
| chr1_-_18118467 | 0.69 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr20_-_38884093 | 0.68 |
ENSDART00000153430
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr21_+_34053590 | 0.68 |
ENSDART00000147519
ENSDART00000158115 ENSDART00000029599 ENSDART00000145123 |
mtmr1b
|
myotubularin related protein 1b |
| chr6_+_28218420 | 0.68 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr3_+_28729443 | 0.64 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr1_-_54570813 | 0.64 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr24_+_8702288 | 0.62 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr20_-_23527234 | 0.61 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr11_+_17849608 | 0.61 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr2_-_57020663 | 0.60 |
|
|
|
| chr16_+_42567707 | 0.60 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr13_-_18564182 | 0.59 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr7_-_24604255 | 0.58 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr11_-_6442490 | 0.58 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr17_+_16038358 | 0.58 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr12_-_33716108 | 0.58 |
ENSDART00000161265
|
peo1
|
progressive external ophthalmoplegia 1 |
| chr6_-_3821922 | 0.56 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr23_-_33692244 | 0.56 |
|
|
|
| chr4_-_75732882 | 0.55 |
|
|
|
| chr16_+_9509875 | 0.54 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr5_-_18502344 | 0.54 |
ENSDART00000137022
ENSDART00000090494 ENSDART00000165701 |
golga3
|
golgin A3 |
| chr5_-_29818204 | 0.54 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr21_+_34053739 | 0.53 |
ENSDART00000147519
|
mtmr1b
|
myotubularin related protein 1b |
| chr10_-_21404605 | 0.52 |
ENSDART00000125167
|
avd
|
avidin |
| chr23_-_35691369 | 0.52 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr16_+_46443800 | 0.52 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr3_-_19890717 | 0.51 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr7_+_17564492 | 0.50 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr14_-_8634381 | 0.50 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr2_+_11335733 | 0.50 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr10_-_35313462 | 0.50 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr1_+_518777 | 0.50 |
ENSDART00000109083
|
txnl4b
|
thioredoxin-like 4B |
| chr17_+_26704439 | 0.49 |
ENSDART00000114927
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr2_-_54224744 | 0.48 |
|
|
|
| chr17_-_25630635 | 0.47 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr7_-_71214600 | 0.46 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr15_-_43954665 | 0.45 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr1_+_23866532 | 0.45 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr10_-_34971926 | 0.45 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr10_-_44713414 | 0.45 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr5_-_26164728 | 0.45 |
ENSDART00000140392
|
rnf181
|
ring finger protein 181 |
| chr14_+_25208174 | 0.44 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr10_+_516924 | 0.44 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr18_+_20571460 | 0.43 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr22_+_8973724 | 0.41 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr1_+_50547385 | 0.41 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr1_+_50547341 | 0.41 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr5_-_4483970 | 0.41 |
|
|
|
| chr18_+_18011759 | 0.41 |
ENSDART00000147797
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr12_+_46281592 | 0.41 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr14_-_33605295 | 0.41 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr10_-_25448712 | 0.41 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr7_+_17564307 | 0.41 |
ENSDART00000158005
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr1_+_18118735 | 0.40 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr13_-_37340209 | 0.40 |
|
|
|
| chr17_-_4086971 | 0.40 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr5_-_29818325 | 0.39 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr12_-_30244575 | 0.39 |
ENSDART00000152981
|
tdrd1
|
tudor domain containing 1 |
| chr15_-_16241412 | 0.39 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr6_+_18941135 | 0.38 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr13_-_42274486 | 0.38 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr24_+_29278310 | 0.38 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr23_-_26241101 | 0.38 |
|
|
|
| chr8_+_26377369 | 0.38 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr25_-_31352866 | 0.38 |
ENSDART00000041740
|
ubl7a
|
ubiquitin-like 7a (bone marrow stromal cell-derived) |
| chr5_-_18502442 | 0.37 |
ENSDART00000165639
|
golga3
|
golgin A3 |
| chr18_+_18011562 | 0.37 |
ENSDART00000005027
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr17_+_12504341 | 0.36 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr5_+_32687626 | 0.36 |
ENSDART00000146759
|
med22
|
mediator complex subunit 22 |
| chr16_+_9509747 | 0.36 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr15_-_16241500 | 0.36 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr17_+_28689850 | 0.36 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| KN149710v1_+_38638 | 0.35 |
|
|
|
| chr17_+_16038103 | 0.35 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr16_+_9509605 | 0.35 |
ENSDART00000162073
ENSDART00000146174 |
ice1
|
KIAA0947-like (H. sapiens) |
| chr19_-_6925090 | 0.35 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr1_+_21244242 | 0.35 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr24_-_14446593 | 0.35 |
|
|
|
| chr1_-_44970931 | 0.35 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr2_+_6341404 | 0.34 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr8_-_21110262 | 0.34 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr12_+_19077980 | 0.34 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr8_+_9955720 | 0.34 |
|
|
|
| chr3_-_15060501 | 0.34 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr9_+_33310981 | 0.33 |
|
|
|
| chr7_-_48393883 | 0.33 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr14_+_15845853 | 0.33 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr3_-_39912816 | 0.33 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr25_-_24440292 | 0.33 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr4_-_20457108 | 0.32 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr21_+_1622040 | 0.32 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr19_-_5186692 | 0.32 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_+_25126238 | 0.32 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr19_+_42657913 | 0.32 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr5_+_26165043 | 0.32 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr19_+_46371377 | 0.32 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr19_+_32614657 | 0.31 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr17_-_43725091 | 0.31 |
|
|
|
| chr14_-_26199859 | 0.31 |
ENSDART00000054175
ENSDART00000145625 |
smad5
|
SMAD family member 5 |
| chr24_+_39630741 | 0.30 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr23_+_32102030 | 0.30 |
ENSDART00000145501
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr12_+_30252424 | 0.30 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr14_+_17092009 | 0.29 |
ENSDART00000129838
|
rnf212
|
ring finger protein 212 |
| chr1_-_22617455 | 0.29 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr18_+_20571513 | 0.29 |
ENSDART00000136710
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr19_-_19806070 | 0.29 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr20_+_38555185 | 0.29 |
ENSDART00000020153
|
adck3
|
aarF domain containing kinase 3 |
| chr1_-_25750237 | 0.29 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr13_-_12257051 | 0.29 |
ENSDART00000124784
|
commd8
|
COMM domain containing 8 |
| chr2_-_37371987 | 0.28 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr8_+_50964745 | 0.28 |
ENSDART00000013870
|
ENSDARG00000007359
|
ENSDARG00000007359 |
| chr18_+_18011800 | 0.28 |
ENSDART00000147797
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr10_+_516885 | 0.28 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr5_+_32687543 | 0.28 |
ENSDART00000123210
|
med22
|
mediator complex subunit 22 |
| chr18_+_20571400 | 0.28 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr8_+_20108592 | 0.27 |
|
|
|
| KN150703v1_+_15225 | 0.27 |
|
|
|
| chr18_+_20571785 | 0.27 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr12_+_3632548 | 0.27 |
|
|
|
| chr23_+_44470297 | 0.26 |
ENSDART00000149842
|
MEPCE
|
methylphosphate capping enzyme |
| chr20_+_54544931 | 0.26 |
|
|
|
| chr24_+_5863597 | 0.26 |
ENSDART00000161104
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr9_-_9729041 | 0.26 |
ENSDART00000175212
|
gpr156
|
G protein-coupled receptor 156 |
| chr21_-_39132951 | 0.26 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr7_-_64173039 | 0.26 |
ENSDART00000172619
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_54811485 | 0.25 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr5_+_6391432 | 0.25 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr10_+_25986401 | 0.25 |
ENSDART00000138516
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr10_+_2656840 | 0.25 |
|
|
|
| chr4_+_5789600 | 0.25 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr18_-_18886144 | 0.24 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr11_-_44738541 | 0.24 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr3_-_34208507 | 0.24 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr3_-_32741894 | 0.24 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr21_+_26689954 | 0.24 |
ENSDART00000065392
|
calm3b
|
calmodulin 3b (phosphorylase kinase, delta) |
| chr22_-_7688508 | 0.24 |
|
|
|
| chr16_+_42567668 | 0.24 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr3_-_26113336 | 0.24 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr7_-_39874079 | 0.24 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr10_-_32550351 | 0.24 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr7_-_17564136 | 0.24 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr3_+_48811662 | 0.23 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr16_-_42105733 | 0.23 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr11_-_33956671 | 0.23 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr20_-_34126039 | 0.23 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr19_+_31998646 | 0.23 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr12_+_46281511 | 0.22 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr20_-_25546095 | 0.22 |
ENSDART00000145034
|
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr12_-_17564384 | 0.22 |
ENSDART00000079065
|
ccz1
|
CCZ1 homolog, vacuolar protein trafficking and biogenesis associated |
| chr2_-_55584106 | 0.22 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr15_+_45738963 | 0.22 |
|
|
|
| chr9_-_35073289 | 0.22 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr6_-_41141242 | 0.22 |
ENSDART00000128723
ENSDART00000151055 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr19_+_770458 | 0.22 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr15_-_44581550 | 0.21 |
|
|
|
| chr15_-_17233876 | 0.21 |
ENSDART00000101718
|
cul5a
|
cullin 5a |
| chr19_+_32317583 | 0.21 |
ENSDART00000151218
|
tpd52
|
tumor protein D52 |
| chr18_+_19467527 | 0.21 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr8_-_21039978 | 0.21 |
ENSDART00000137606
ENSDART00000146532 |
zgc:112962
|
zgc:112962 |
| chr13_+_36797201 | 0.21 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr18_-_43890836 | 0.21 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr11_-_44539726 | 0.21 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr8_-_4562273 | 0.20 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr9_-_2522639 | 0.20 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr6_-_41141280 | 0.20 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr16_-_42105636 | 0.20 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr4_-_14894468 | 0.20 |
|
|
|
| chr7_-_54752916 | 0.19 |
|
|
|
| chr12_+_46281623 | 0.19 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr2_-_33703921 | 0.19 |
ENSDART00000147439
|
atp6v0b
|
ATPase, H+ transporting, lysosomal V0 subunit b |
| chr21_-_30957754 | 0.19 |
ENSDART00000065503
|
pgap2
|
post-GPI attachment to proteins 2 |
| chr12_+_19077847 | 0.19 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr5_-_3103278 | 0.19 |
|
|
|
| chr22_-_3986398 | 0.19 |
ENSDART00000169317
ENSDART00000171262 |
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr24_+_23571714 | 0.19 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr24_+_34183407 | 0.19 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr3_-_26112995 | 0.19 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr10_+_2554914 | 0.18 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin-like 2 |
| chr2_+_6341345 | 0.18 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr11_+_17849528 | 0.18 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr25_-_36691657 | 0.18 |
ENSDART00000153789
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr21_+_15815265 | 0.18 |
ENSDART00000154468
|
AL953866.1
|
ENSDARG00000096884 |
| chr2_+_10209233 | 0.18 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr8_+_28362546 | 0.18 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr21_-_25765319 | 0.17 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr22_+_2074020 | 0.17 |
ENSDART00000106540
|
znf1161
|
zinc finger protein 1161 |
| chr8_+_39640881 | 0.17 |
ENSDART00000040330
ENSDART00000045529 ENSDART00000159852 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr7_-_10318692 | 0.17 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.3 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 0.8 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.6 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.4 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0009223 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.8 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.1 | 0.4 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.0 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.3 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.6 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 0.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.3 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.5 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.9 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.6 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.6 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.2 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.2 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.1 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0000215 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.4 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |