Project
DANIO-CODE
Navigation
Downloads
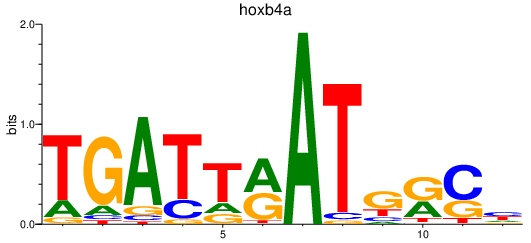
Results for hoxb4a
Z-value: 0.26
Transcription factors associated with hoxb4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb4a
|
ENSDARG00000013533 | homeobox B4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb4a | dr10_dc_chr3_+_23590779_23590782 | 0.42 | 1.1e-01 | Click! |
Activity profile of hoxb4a motif
Sorted Z-values of hoxb4a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb4a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_14197118 | 0.42 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr21_-_20291707 | 0.34 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr20_-_29517770 | 0.32 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr22_-_22337268 | 0.30 |
|
|
|
| chr6_+_29800606 | 0.30 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr6_+_4712785 | 0.29 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr3_+_26896869 | 0.27 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr12_+_28739504 | 0.26 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr23_+_27830375 | 0.23 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr19_+_20209561 | 0.23 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr17_+_27417635 | 0.21 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr22_+_11393451 | 0.20 |
|
|
|
| chr4_-_19027117 | 0.20 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr3_+_33168814 | 0.19 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr15_-_31687 | 0.19 |
|
|
|
| chr5_+_37113743 | 0.18 |
|
|
|
| chr17_-_19606453 | 0.18 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr10_-_26782374 | 0.17 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr8_+_28419432 | 0.16 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr23_+_14058972 | 0.15 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr21_+_22386643 | 0.15 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr8_+_22910147 | 0.14 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr12_+_9664920 | 0.14 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr5_+_19608261 | 0.12 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr13_+_30781652 | 0.12 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr19_+_816354 | 0.11 |
ENSDART00000093304
|
nrm
|
nurim (nuclear envelope membrane protein |
| chr12_-_26339002 | 0.11 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr4_-_16461748 | 0.11 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr16_-_19154920 | 0.10 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr5_-_63422783 | 0.09 |
ENSDART00000083684
|
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr21_-_27301938 | 0.09 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr7_-_20201358 | 0.08 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr7_-_20201693 | 0.08 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr13_+_30781718 | 0.08 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr9_+_32267615 | 0.08 |
|
|
|
| chr16_+_21122024 | 0.07 |
|
|
|
| chr23_+_27830103 | 0.07 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr3_+_35277555 | 0.07 |
|
|
|
| chr19_+_14197020 | 0.06 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr5_-_7329952 | 0.06 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr17_-_8570443 | 0.05 |
ENSDART00000049236
|
ctbp2a
|
C-terminal binding protein 2a |
| chr3_+_24007046 | 0.04 |
|
|
|
| chr23_+_27830256 | 0.04 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr10_+_36082726 | 0.04 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr17_-_8570497 | 0.03 |
ENSDART00000148827
|
ctbp2a
|
C-terminal binding protein 2a |
| chr1_-_686007 | 0.03 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_+_23621561 | 0.02 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr9_+_32267297 | 0.02 |
|
|
|
| chr18_+_13589341 | 0.02 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr18_+_5986137 | 0.01 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr2_+_54075496 | 0.01 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr1_+_1564283 | 0.00 |
ENSDART00000166968
|
atp1a1a.2
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 2 |
| chr4_-_2707563 | 0.00 |
ENSDART00000021953
ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |