Project
DANIO-CODE
Navigation
Downloads
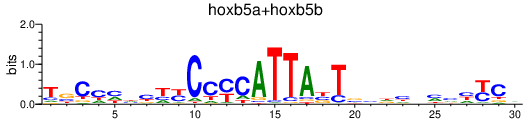
Results for hoxb5a+hoxb5b
Z-value: 0.35
Transcription factors associated with hoxb5a+hoxb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb5a
|
ENSDARG00000013057 | homeobox B5a |
|
hoxb5b
|
ENSDARG00000054030 | homeobox B5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb5b | dr10_dc_chr12_+_27038298_27038327 | 0.20 | 4.6e-01 | Click! |
| hoxb5a | dr10_dc_chr3_+_23577103_23577162 | 0.10 | 7.2e-01 | Click! |
Activity profile of hoxb5a+hoxb5b motif
Sorted Z-values of hoxb5a+hoxb5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb5a+hoxb5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| KN149799v1_-_24611 | 1.31 |
|
|
|
| KN149799v1_+_25349 | 0.37 |
|
|
|
| chr8_-_17692185 | 0.23 |
|
|
|
| chr16_+_53438473 | 0.21 |
ENSDART00000155940
|
CR848746.1
|
ENSDARG00000097164 |
| chr9_+_16846294 | 0.20 |
ENSDART00000110866
|
cln5
|
ceroid-lipofuscinosis, neuronal 5 |
| chr19_+_43745515 | 0.19 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr9_-_12063411 | 0.18 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr6_+_37881015 | 0.17 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr15_-_42327430 | 0.17 |
ENSDART00000144645
|
farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr5_+_31745031 | 0.15 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr19_+_5562107 | 0.15 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr16_+_53438411 | 0.14 |
ENSDART00000155940
|
CR848746.1
|
ENSDARG00000097164 |
| chr13_-_36008642 | 0.12 |
ENSDART00000015664
|
pcnx
|
pecanex homolog (Drosophila) |
| chr22_+_659157 | 0.11 |
ENSDART00000149712
|
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr18_-_616875 | 0.11 |
ENSDART00000169974
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr23_-_624480 | 0.11 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr8_-_17691889 | 0.11 |
|
|
|
| chr23_-_624534 | 0.11 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr2_-_5233961 | 0.11 |
ENSDART00000163728
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr7_+_50828247 | 0.10 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr23_+_20592121 | 0.10 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr24_-_42193714 | 0.09 |
|
|
|
| chr20_+_51918693 | 0.09 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr7_-_50827308 | 0.09 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr14_+_33382973 | 0.08 |
ENSDART00000132488
|
apln
|
apelin |
| chr15_-_723851 | 0.08 |
|
|
|
| chr24_+_42193701 | 0.08 |
ENSDART00000067734
|
snx16
|
sorting nexin 16 |
| chr3_+_39437497 | 0.08 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr22_+_1663690 | 0.08 |
ENSDART00000166672
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr8_+_166461 | 0.08 |
|
|
|
| chr12_-_4638371 | 0.07 |
|
|
|
| chr7_+_50828119 | 0.07 |
ENSDART00000108738
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr23_+_3570270 | 0.07 |
ENSDART00000092258
|
spata2
|
spermatogenesis associated 2 |
| chr13_-_36008571 | 0.07 |
ENSDART00000139593
|
pcnx
|
pecanex homolog (Drosophila) |
| chr3_+_28700619 | 0.06 |
ENSDART00000108973
|
flr
|
fleer |
| chr10_+_26982647 | 0.06 |
ENSDART00000135493
|
frmd8
|
FERM domain containing 8 |
| chr19_+_43745687 | 0.06 |
ENSDART00000102384
|
sesn2
|
sestrin 2 |
| chr9_-_32295031 | 0.06 |
ENSDART00000132448
|
BX530407.4
|
ENSDARG00000093950 |
| chr14_+_27936303 | 0.06 |
ENSDART00000003293
|
mid2
|
midline 2 |
| chr2_-_5233883 | 0.06 |
ENSDART00000163728
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr5_-_33169062 | 0.06 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| KN149874v1_-_3544 | 0.06 |
ENSDART00000165981
|
CABZ01046348.1
|
ENSDARG00000099481 |
| chr19_+_46667326 | 0.05 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr16_-_42202425 | 0.05 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr7_+_33044157 | 0.05 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr19_-_23259063 | 0.05 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| KN149874v1_+_3898 | 0.05 |
|
|
|
| KN150226v1_+_11579 | 0.04 |
|
|
|
| chr2_+_24661778 | 0.04 |
ENSDART00000139339
|
mrpl34
|
mitochondrial ribosomal protein L34 |
| chr11_+_24720206 | 0.04 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr5_+_33807197 | 0.04 |
ENSDART00000145127
|
lamc3
|
laminin, gamma 3 |
| chr3_+_1455242 | 0.03 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr1_+_54403123 | 0.03 |
ENSDART00000152654
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr11_+_24720057 | 0.03 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr15_-_25164321 | 0.03 |
ENSDART00000154628
|
fam101b
|
family with sequence similarity 101, member B |
| chr23_+_35980812 | 0.03 |
|
|
|
| chr19_+_43745546 | 0.03 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr3_-_2041373 | 0.02 |
ENSDART00000146552
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr19_+_31046291 | 0.02 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr16_-_10089440 | 0.02 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr19_+_40535249 | 0.01 |
ENSDART00000151276
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr17_+_20549925 | 0.01 |
ENSDART00000113936
|
ENSDARG00000074663
|
ENSDARG00000074663 |
| chr24_+_37805069 | 0.01 |
|
|
|
| chr5_-_29480173 | 0.01 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr1_-_9316038 | 0.01 |
|
|
|
| chr23_+_3570243 | 0.01 |
ENSDART00000092258
|
spata2
|
spermatogenesis associated 2 |
| chr12_+_46703637 | 0.00 |
ENSDART00000152928
|
exoc7
|
exocyst complex component 7 |
| chr14_-_8779307 | 0.00 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| KN150563v1_-_10770 | 0.00 |
|
|
|
| chr9_-_1954910 | 0.00 |
ENSDART00000140438
|
BX322661.1
|
ENSDARG00000094186 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.2 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0060544 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |