Project
DANIO-CODE
Navigation
Downloads
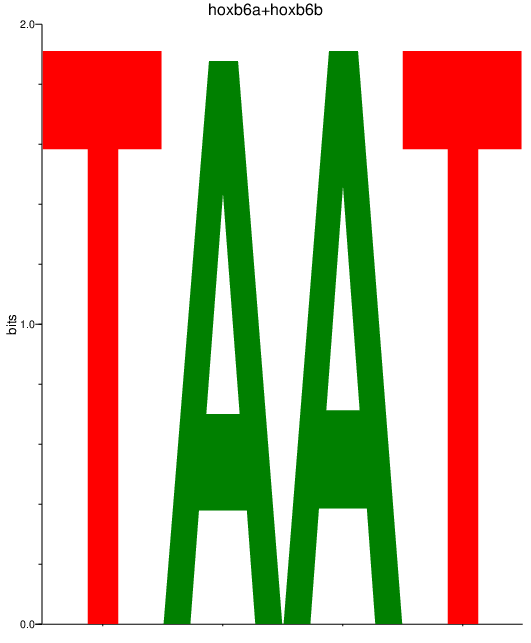
Results for hoxb6a+hoxb6b
Z-value: 4.02
Transcription factors associated with hoxb6a+hoxb6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb6a
|
ENSDARG00000010630 | homeobox B6a |
|
hoxb6b
|
ENSDARG00000026513 | homeobox B6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb6b | dr10_dc_chr12_+_27035744_27035785 | 0.95 | 1.7e-08 | Click! |
| hoxb6a | dr10_dc_chr3_+_23573053_23573058 | 0.87 | 1.4e-05 | Click! |
Activity profile of hoxb6a+hoxb6b motif
Sorted Z-values of hoxb6a+hoxb6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb6a+hoxb6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32686790 | 9.96 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr21_+_7844259 | 7.89 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr1_+_9367672 | 7.54 |
ENSDART00000144756
|
fgb
|
fibrinogen beta chain |
| chr16_+_24069711 | 6.97 |
ENSDART00000142869
|
apoc2
|
apolipoprotein C-II |
| chr20_-_22576513 | 6.81 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr24_-_1307905 | 6.80 |
ENSDART00000159212
|
nrp1a
|
neuropilin 1a |
| chr22_-_35967525 | 6.80 |
ENSDART00000176971
|
CABZ01037120.1
|
ENSDARG00000106725 |
| chr10_+_39141022 | 6.75 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr4_+_5498355 | 6.58 |
ENSDART00000150785
|
mapk11
|
mitogen-activated protein kinase 11 |
| chr12_+_5673442 | 6.39 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr15_+_32853646 | 6.29 |
ENSDART00000167515
|
postnb
|
periostin, osteoblast specific factor b |
| chr4_-_15442828 | 6.21 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr1_+_43738350 | 5.91 |
ENSDART00000127087
ENSDART00000100309 |
crybb1l2
|
crystallin, beta B1, like 2 |
| chr15_+_28753020 | 5.88 |
ENSDART00000155815
|
nova2
|
neuro-oncological ventral antigen 2 |
| chr19_+_7234029 | 5.78 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr9_-_32942783 | 5.70 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr13_+_23065500 | 5.65 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr11_-_44724371 | 5.64 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr19_-_41887040 | 5.53 |
ENSDART00000062026
|
dlx5a
|
distal-less homeobox 5a |
| chr24_-_28814066 | 5.52 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr16_+_46145286 | 5.51 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr25_+_30827761 | 5.39 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr2_-_28446615 | 5.34 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr21_-_8420626 | 5.31 |
ENSDART00000084378
|
crb2a
|
crumbs family member 2a |
| chr12_+_9664920 | 5.27 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr23_+_35964754 | 5.20 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr1_-_38097100 | 5.18 |
ENSDART00000148572
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr18_+_38773971 | 5.18 |
ENSDART00000010177
|
onecut1
|
one cut homeobox 1 |
| chr2_+_24649130 | 5.10 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr8_+_23464087 | 5.09 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr13_+_35213326 | 5.08 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr7_+_30516734 | 5.07 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr22_+_15995914 | 5.05 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr1_+_6862901 | 5.05 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr15_-_18425091 | 5.02 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr1_-_4757890 | 5.01 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr11_+_21749658 | 4.96 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr22_-_32553488 | 4.91 |
ENSDART00000104693
|
pcbp4
|
poly(rC) binding protein 4 |
| chr25_+_21732255 | 4.89 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr7_-_55406347 | 4.89 |
ENSDART00000021009
|
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr12_-_34614931 | 4.85 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr16_+_46327528 | 4.82 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr5_+_49093134 | 4.78 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr15_-_24987099 | 4.76 |
|
|
|
| chr23_-_31419805 | 4.75 |
ENSDART00000138106
|
phip
|
pleckstrin homology domain interacting protein |
| chr18_+_9213713 | 4.73 |
ENSDART00000127469
ENSDART00000101192 |
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr13_+_28997608 | 4.70 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr2_-_34010299 | 4.68 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr22_-_16015935 | 4.60 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr9_-_1954910 | 4.57 |
ENSDART00000140438
|
BX322661.1
|
ENSDARG00000094186 |
| chr16_+_7213161 | 4.54 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr7_-_25623974 | 4.53 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr9_+_37157463 | 4.52 |
ENSDART00000059753
|
inhbb
|
inhibin, beta B |
| chr16_+_16941228 | 4.51 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr21_-_35806638 | 4.45 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr19_-_2486568 | 4.44 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr19_+_7233537 | 4.43 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr16_+_29716279 | 4.40 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr23_+_6861489 | 4.39 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr5_+_49093250 | 4.39 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr22_+_11726312 | 4.38 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr5_-_71643185 | 4.37 |
ENSDART00000029014
|
pax8
|
paired box 8 |
| chr1_+_26330186 | 4.37 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr17_+_3879416 | 4.33 |
ENSDART00000151849
|
hao1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr4_-_2615160 | 4.32 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr20_-_9107294 | 4.32 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr17_-_14868764 | 4.31 |
ENSDART00000115064
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr24_-_23175007 | 4.29 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr21_+_28408329 | 4.27 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr8_-_38126693 | 4.26 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr9_-_13383818 | 4.26 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr1_-_25600988 | 4.25 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr6_+_29226868 | 4.23 |
ENSDART00000006386
|
atp1b1a
|
ATPase, Na+/K+ transporting, beta 1a polypeptide |
| chr1_+_14540517 | 4.22 |
|
|
|
| chr24_+_17125429 | 4.21 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr5_+_44316830 | 4.20 |
|
|
|
| chr4_-_11752178 | 4.18 |
ENSDART00000102301
|
podxl
|
podocalyxin-like |
| chr2_-_20941256 | 4.17 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr20_-_38714872 | 4.15 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr24_-_33869817 | 4.15 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr1_+_13779755 | 4.14 |
|
|
|
| chr22_-_4264838 | 4.13 |
ENSDART00000125302
|
fbn2b
|
fibrillin 2b |
| chr10_-_24422257 | 4.10 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr8_-_33372638 | 4.10 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| chr11_+_8558222 | 4.09 |
ENSDART00000169141
ENSDART00000126523 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr23_-_7740845 | 4.08 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr8_-_49443058 | 4.07 |
ENSDART00000011453
|
sypb
|
synaptophysin b |
| chr7_-_38590391 | 4.07 |
ENSDART00000037361
ENSDART00000173629 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr17_-_36988937 | 4.07 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr22_-_15567180 | 4.06 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr17_+_33766838 | 4.06 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr19_-_6384776 | 4.05 |
|
|
|
| chr14_-_4166292 | 4.02 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr6_+_16279737 | 4.00 |
ENSDART00000040035
|
ccdc80l1
|
coiled-coil domain containing 80 like 1 |
| chr10_+_15819127 | 4.00 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr13_+_28997676 | 3.99 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr17_+_24668907 | 3.98 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr5_-_71539823 | 3.98 |
ENSDART00000167872
|
tbx3a
|
T-box 3a |
| chr16_+_14817799 | 3.96 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr16_+_7213011 | 3.96 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr6_+_56157608 | 3.96 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr24_-_21778717 | 3.95 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr22_+_16509286 | 3.95 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr25_-_7635824 | 3.93 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr16_-_24220413 | 3.92 |
ENSDART00000103176
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr8_-_34077387 | 3.91 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr13_+_27102308 | 3.90 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr19_-_35739239 | 3.90 |
|
|
|
| chr8_-_18502159 | 3.89 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr6_+_52791515 | 3.89 |
ENSDART00000065682
|
matn4
|
matrilin 4 |
| chr10_+_18994733 | 3.88 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr14_-_42633578 | 3.88 |
ENSDART00000162714
ENSDART00000161521 |
pcdh10b
|
protocadherin 10b |
| chr1_-_24227846 | 3.86 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr22_-_15930756 | 3.86 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr16_+_38409924 | 3.84 |
ENSDART00000087346
|
zgc:113232
|
zgc:113232 |
| chr2_-_23516930 | 3.84 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr5_-_14148028 | 3.83 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr12_+_24221087 | 3.82 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr12_-_4767887 | 3.79 |
ENSDART00000167490
|
mapta
|
microtubule-associated protein tau a |
| chr2_-_58111727 | 3.79 |
ENSDART00000004431
ENSDART00000166845 ENSDART00000163999 |
epb41l3b
|
erythrocyte membrane protein band 4.1-like 3b |
| chr23_+_35996491 | 3.75 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr3_+_51182697 | 3.75 |
|
|
|
| chr12_-_47699958 | 3.74 |
ENSDART00000171932
ENSDART00000168165 ENSDART00000161985 |
HHEX
|
hematopoietically expressed homeobox |
| chr19_-_6856033 | 3.74 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr16_+_26900732 | 3.74 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr20_-_45908435 | 3.69 |
ENSDART00000147637
|
fermt1
|
fermitin family member 1 |
| chr20_+_47588954 | 3.68 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr2_-_18275994 | 3.67 |
ENSDART00000155124
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr14_-_2008649 | 3.65 |
ENSDART00000161817
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr19_+_12058673 | 3.63 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr1_+_50835265 | 3.63 |
ENSDART00000162226
|
meis1a
|
Meis homeobox 1 a |
| chr17_-_32473712 | 3.62 |
ENSDART00000148455
ENSDART00000149885 |
grhl1
|
grainyhead-like transcription factor 1 |
| chr23_-_31446156 | 3.61 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr8_+_15989815 | 3.61 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr25_+_19655928 | 3.60 |
ENSDART00000128852
|
celsr1b
|
cadherin EGF LAG seven-pass G-type receptor 1b |
| chr16_+_5256773 | 3.60 |
ENSDART00000012053
|
elovl4a
|
ELOVL fatty acid elongase 4a |
| chr14_-_2682064 | 3.60 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr25_+_18467217 | 3.60 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr13_+_27102377 | 3.59 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr11_-_38843219 | 3.58 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr19_-_18316262 | 3.58 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr5_-_47523737 | 3.57 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr11_-_29910947 | 3.57 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr9_+_12988528 | 3.55 |
ENSDART00000125961
|
bmpr2b
|
bone morphogenetic protein receptor, type II b (serine/threonine kinase) |
| chr23_+_43362722 | 3.55 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr13_+_33151628 | 3.52 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr16_-_39620777 | 3.52 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr13_+_33378267 | 3.50 |
ENSDART00000025007
|
jag2a
|
jagged 2a |
| chr1_-_44476084 | 3.50 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr11_+_14142126 | 3.50 |
ENSDART00000102520
|
palm1a
|
paralemmin 1a |
| chr9_+_41720118 | 3.50 |
|
|
|
| chr18_-_14709371 | 3.50 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr19_-_22182031 | 3.49 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr16_-_29593569 | 3.48 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr19_-_47994946 | 3.47 |
ENSDART00000114549
|
CU695215.1
|
ENSDARG00000076126 |
| chr3_+_23546802 | 3.47 |
ENSDART00000023674
|
hoxb9a
|
homeobox B9a |
| chr16_+_49796978 | 3.46 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr24_+_24308055 | 3.46 |
|
|
|
| chr5_-_47656197 | 3.45 |
|
|
|
| chr5_-_40133824 | 3.45 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr23_+_36023748 | 3.45 |
|
|
|
| chr24_-_6048914 | 3.44 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr19_+_41488385 | 3.43 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr6_+_42821679 | 3.43 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr16_-_24697750 | 3.43 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr13_-_31491759 | 3.42 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr20_+_33391554 | 3.41 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr7_+_15623852 | 3.41 |
ENSDART00000161608
|
pax6b
|
paired box 6b |
| chr6_+_6767424 | 3.40 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr20_-_42805703 | 3.40 |
ENSDART00000045816
|
plg
|
plasminogen |
| chr20_-_29517770 | 3.39 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr1_-_29706856 | 3.39 |
ENSDART00000174868
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr13_-_33696425 | 3.39 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr7_+_25794014 | 3.38 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr2_-_17564676 | 3.38 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr6_-_8001756 | 3.35 |
ENSDART00000151483
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr9_+_31471391 | 3.35 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr12_+_25684420 | 3.35 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr14_+_35880163 | 3.34 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr4_+_12932838 | 3.34 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr16_+_43249142 | 3.33 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr6_-_16590883 | 3.33 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr5_-_28025315 | 3.33 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr23_+_17856053 | 3.32 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr13_+_22528640 | 3.31 |
ENSDART00000078877
|
sncga
|
synuclein, gamma a |
| chr11_-_10675245 | 3.31 |
ENSDART00000115255
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr13_-_36265504 | 3.31 |
ENSDART00000140243
|
actn1
|
actinin, alpha 1 |
| chr24_-_23797468 | 3.29 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr25_+_18467835 | 3.28 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr16_-_24634751 | 3.28 |
ENSDART00000108590
|
si:ch211-79k12.1
|
si:ch211-79k12.1 |
| chr14_+_11151485 | 3.28 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr3_+_28808901 | 3.27 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr6_-_50705420 | 3.27 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr12_-_25825072 | 3.26 |
|
|
|
| chr11_+_36215554 | 3.23 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr20_+_16843502 | 3.22 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr14_-_11150618 | 3.22 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr13_+_11305781 | 3.21 |
|
|
|
| chr25_+_34684585 | 3.21 |
ENSDART00000044453
|
ano5a
|
anoctamin 5a |
| chr3_+_24327586 | 3.19 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.8 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 2.9 | 8.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 2.8 | 11.3 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 2.3 | 6.8 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 2.1 | 8.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 2.0 | 8.0 | GO:0086026 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 2.0 | 5.9 | GO:0033632 | regulation of cell adhesion mediated by integrin(GO:0033628) positive regulation of cell adhesion mediated by integrin(GO:0033630) cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.9 | 7.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.9 | 7.7 | GO:0098900 | regulation of action potential(GO:0098900) |
| 1.9 | 5.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 1.7 | 5.2 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 1.7 | 5.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 1.7 | 5.0 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 1.6 | 6.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.5 | 5.9 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 1.5 | 14.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 1.4 | 4.3 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 1.4 | 4.3 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 1.4 | 4.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 1.4 | 4.3 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 1.4 | 7.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 1.4 | 4.2 | GO:0030431 | sleep(GO:0030431) |
| 1.4 | 4.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 1.3 | 3.9 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 1.3 | 3.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 1.3 | 5.0 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 1.3 | 5.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 1.2 | 6.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 1.2 | 4.9 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 1.2 | 3.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 1.2 | 13.9 | GO:0021767 | mammillary body development(GO:0021767) |
| 1.2 | 2.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 1.1 | 9.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 1.1 | 12.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 1.1 | 3.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 1.1 | 3.4 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 1.1 | 3.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 1.1 | 4.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.1 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 1.1 | 6.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.0 | 6.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.0 | 3.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 2.0 | GO:0061195 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 1.0 | 7.0 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.0 | 2.0 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 1.0 | 6.8 | GO:0031641 | regulation of myelination(GO:0031641) |
| 1.0 | 8.8 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) pattern specification involved in pronephros development(GO:0039017) pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 1.0 | 3.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.9 | 4.7 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.9 | 1.8 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.9 | 5.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.9 | 14.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.9 | 2.7 | GO:0051701 | interaction with host(GO:0051701) |
| 0.9 | 2.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.9 | 9.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.8 | 2.5 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.8 | 5.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.8 | 2.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.8 | 4.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.8 | 3.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.8 | 5.5 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.8 | 6.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.7 | 4.4 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.7 | 1.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.7 | 3.6 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.7 | 2.2 | GO:0022029 | telencephalon cell migration(GO:0022029) |
| 0.7 | 2.1 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.7 | 6.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.7 | 2.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.7 | 4.9 | GO:0021779 | oligodendrocyte cell fate commitment(GO:0021779) glial cell fate commitment(GO:0021781) |
| 0.7 | 2.1 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.7 | 1.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.7 | 2.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.7 | 0.7 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.7 | 3.4 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.7 | 19.1 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.7 | 2.7 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.7 | 2.7 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.7 | 2.7 | GO:0021561 | facial nerve development(GO:0021561) rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.7 | 2.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.7 | 11.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.7 | 2.6 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.7 | 2.6 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.6 | 9.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.6 | 8.3 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.6 | 1.3 | GO:0042330 | taxis(GO:0042330) |
| 0.6 | 1.9 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.6 | 2.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.6 | 3.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.6 | 1.8 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.6 | 2.4 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.6 | 3.6 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.6 | 2.4 | GO:0071387 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.6 | 3.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.6 | 4.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.6 | 4.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.6 | 1.7 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.6 | 4.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.5 | 1.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.5 | 1.6 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.5 | 6.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.5 | 7.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.5 | 2.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.5 | 2.1 | GO:0050848 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) regulation of calcium-mediated signaling(GO:0050848) |
| 0.5 | 2.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.5 | 3.1 | GO:0032371 | membrane raft organization(GO:0031579) regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.5 | 1.6 | GO:0006014 | D-ribose metabolic process(GO:0006014) pentose catabolic process(GO:0019323) |
| 0.5 | 1.5 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.5 | 2.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.5 | 2.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.5 | 5.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.5 | 1.5 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) chondrocyte morphogenesis(GO:0090171) |
| 0.5 | 6.4 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.5 | 2.9 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.5 | 3.9 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.5 | 2.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.5 | 6.7 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.5 | 1.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.5 | 9.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.5 | 1.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.5 | 3.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.5 | 1.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.5 | 2.3 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.4 | 2.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.4 | 2.2 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.4 | 4.0 | GO:0046323 | glucose import(GO:0046323) |
| 0.4 | 1.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 1.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.4 | 1.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 2.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.4 | 2.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.4 | 1.3 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.4 | 0.4 | GO:0044539 | medium-chain fatty acid transport(GO:0001579) long-chain fatty acid import(GO:0044539) |
| 0.4 | 2.5 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.4 | 1.6 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.4 | 1.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.4 | 1.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.4 | 2.4 | GO:0045109 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 1.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.4 | 2.4 | GO:0070293 | renal absorption(GO:0070293) |
| 0.4 | 2.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 3.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.4 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 2.4 | GO:0001964 | startle response(GO:0001964) |
| 0.4 | 0.8 | GO:0050864 | regulation of B cell activation(GO:0050864) |
| 0.4 | 3.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 2.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.4 | 1.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.4 | 2.6 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.4 | 4.1 | GO:0060324 | face development(GO:0060324) |
| 0.4 | 1.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.4 | 22.4 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.4 | 0.7 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.4 | 1.9 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.4 | 2.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.4 | 1.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.4 | 2.9 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.4 | 2.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.4 | 2.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.4 | 4.3 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.4 | 5.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 1.8 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.4 | 2.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 1.4 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 2.4 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.3 | 2.4 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.3 | 1.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 1.0 | GO:0014005 | microglia development(GO:0014005) |
| 0.3 | 2.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 1.7 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 6.0 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.3 | 2.0 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.3 | 2.7 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.3 | 1.3 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.3 | 5.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 2.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 1.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.3 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 1.3 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.3 | 1.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 2.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.3 | 1.0 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 3.5 | GO:0032946 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.3 | 4.5 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.3 | 1.3 | GO:0032355 | glutathione catabolic process(GO:0006751) response to estradiol(GO:0032355) |
| 0.3 | 3.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.3 | 5.6 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.3 | 0.6 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 0.3 | 3.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 2.7 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.3 | 28.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 1.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 2.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 7.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.3 | 1.7 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.3 | 4.0 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.3 | 2.3 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.3 | 0.9 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.3 | 0.6 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 | 1.1 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 0.3 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.3 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 1.4 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.3 | 1.9 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.3 | 2.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.3 | 1.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 4.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 | 6.8 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.3 | 3.4 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.3 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 4.4 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.3 | 1.6 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.3 | 4.7 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.3 | 1.5 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.3 | 1.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.3 | 0.8 | GO:0060996 | dendritic spine development(GO:0060996) dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 0.8 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.2 | 2.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 5.7 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.2 | 0.7 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 9.8 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.2 | 1.4 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 5.4 | GO:2000021 | regulation of ion homeostasis(GO:2000021) |
| 0.2 | 0.7 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.2 | 1.4 | GO:0060021 | palate development(GO:0060021) |
| 0.2 | 1.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 4.2 | GO:0021761 | limbic system development(GO:0021761) |
| 0.2 | 0.9 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.2 | 1.6 | GO:1901906 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 2.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 1.3 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.2 | 1.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.2 | 4.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 1.3 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 4.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.2 | 2.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 2.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 0.4 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.2 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 5.6 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.2 | 1.6 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 20.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.2 | 0.4 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.2 | 1.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 1.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.2 | 4.0 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.2 | 3.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.2 | 5.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 3.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 3.3 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.2 | 4.1 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.2 | 1.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 1.0 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 3.5 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.2 | 1.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 8.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.2 | 0.6 | GO:0035872 | peptidoglycan transport(GO:0015835) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.2 | 2.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 1.3 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 1.5 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.2 | 4.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 1.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 3.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 0.9 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.2 | 0.5 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 3.4 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.2 | 2.8 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.2 | 1.8 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.2 | 1.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.8 | GO:0048941 | lateral line nerve glial cell differentiation(GO:0048895) myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell development(GO:0048937) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.2 | 9.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.2 | 2.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 2.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.2 | 13.6 | GO:0099536 | synaptic signaling(GO:0099536) |
| 0.2 | 0.6 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.4 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.3 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 1.3 | GO:0051647 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) nucleus localization(GO:0051647) |
| 0.1 | 0.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 9.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.1 | 1.7 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.1 | 2.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 9.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 3.7 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.1 | 5.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.5 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 3.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 7.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 2.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 2.0 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.3 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 2.2 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 2.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 7.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.3 | GO:0002548 | monocyte chemotaxis(GO:0002548) eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 1.2 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 2.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.2 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.8 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 4.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.0 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.8 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.6 | GO:0071248 | cellular response to metal ion(GO:0071248) |
| 0.1 | 1.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0048919 | posterior lateral line neuromast development(GO:0048919) |
| 0.1 | 1.7 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.0 | GO:0048753 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.1 | 4.4 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 4.4 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.9 | GO:1902110 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 1.0 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 3.5 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.1 | 0.3 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 1.2 | GO:0061572 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 2.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.2 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.2 | GO:0007612 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 3.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 2.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.0 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 1.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 1.3 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.7 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.1 | 1.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.5 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.8 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.1 | 0.2 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 0.4 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0072422 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0030804 | positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) positive regulation of cyclase activity(GO:0031281) regulation of adenylate cyclase activity(GO:0045761) positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.0 | 3.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.5 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.9 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.8 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.3 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.6 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.6 | GO:0030203 | glycosaminoglycan metabolic process(GO:0030203) |
| 0.0 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.3 | GO:1903051 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 | 0.3 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 1.2 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.8 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.4 | 7.0 | GO:0042627 | chylomicron(GO:0042627) |
| 1.2 | 6.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.2 | 1.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.0 | 7.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 1.0 | 5.9 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.8 | 6.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.8 | 6.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.7 | 4.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.7 | 4.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.6 | 1.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.6 | 1.8 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.6 | 1.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.5 | 2.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 11.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.4 | 15.2 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.4 | 2.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 11.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 1.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 1.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 1.4 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 2.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 1.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 1.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 7.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 0.9 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.3 | 5.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.3 | 11.3 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.3 | 6.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 3.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 1.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 4.9 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.3 | 11.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 1.0 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 47.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 1.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 1.5 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 16.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.2 | 2.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 2.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 2.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 2.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.2 | 13.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 5.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 0.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 15.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 3.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 2.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 9.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 2.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 31.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 1.8 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 7.0 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.2 | 1.2 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 10.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 1.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 2.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 3.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 6.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 6.2 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 0.4 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 1.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.6 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.1 | 62.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 3.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 9.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.1 | 1.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.8 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 24.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 1.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.1 | 0.3 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 5.2 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 2.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.3 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 2.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 1.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 9.3 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 18.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 1.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.4 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 4.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 45.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 10.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.4 | 7.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.3 | 6.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.3 | 5.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.2 | 6.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.2 | 11.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.2 | 3.5 | GO:0034713 | transforming growth factor beta receptor activity, type II(GO:0005026) type I transforming growth factor beta receptor binding(GO:0034713) |
| 1.2 | 4.7 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 1.1 | 3.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.1 | 8.8 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.1 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 1.1 | 6.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.1 | 3.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 1.0 | 6.8 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 1.0 | 7.7 | GO:0005113 | patched binding(GO:0005113) |
| 1.0 | 3.8 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.9 | 3.6 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.9 | 4.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.8 | 3.4 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.8 | 11.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.8 | 11.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.8 | 2.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.8 | 2.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.8 | 2.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.8 | 7.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.8 | 7.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.8 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.8 | 2.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.8 | 2.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.7 | 4.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.7 | 2.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.7 | 10.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.7 | 2.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.7 | 2.6 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.6 | 2.5 | GO:0035620 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.6 | 1.8 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.6 | 1.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.6 | 1.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.6 | 3.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.6 | 2.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.6 | 1.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 8.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.5 | 14.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 5.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.5 | 2.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 9.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.5 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 1.6 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.5 | 11.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 3.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 5.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.5 | 2.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.5 | 8.3 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.5 | 1.8 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.4 | 2.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.4 | 5.2 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.4 | 4.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 1.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 5.9 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.4 | 1.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.4 | 4.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.4 | 23.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 2.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.4 | 1.6 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.4 | 2.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 1.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 1.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 6.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 1.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 1.8 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.4 | 2.1 | GO:0004931 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 3.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 4.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 1.0 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.3 | 2.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 24.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 2.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 1.9 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.3 | 4.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 9.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.6 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.3 | 2.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 5.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 0.9 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.3 | 5.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 6.1 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.3 | 4.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 2.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.3 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 2.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 9.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 1.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 14.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.3 | 0.8 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.3 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 3.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 1.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 5.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 1.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.0 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 0.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 2.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 11.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 0.9 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 6.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.6 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 1.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 3.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.2 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 0.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 2.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 26.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 2.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 2.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 12.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.2 | 2.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 0.6 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 0.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 5.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 1.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 7.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 240.7 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.2 | 10.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 0.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 5.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 1.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 5.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 5.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.2 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 0.8 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.8 | GO:0016933 | excitatory extracellular ligand-gated ion channel activity(GO:0005231) inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 10.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 13.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 3.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 10.0 | GO:0004888 | transmembrane signaling receptor activity(GO:0004888) |
| 0.1 | 1.0 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.1 | 0.5 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 3.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.3 | GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.1 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.5 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 0.4 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 3.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.9 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 37.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 2.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 1.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 1.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 12.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.3 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 1.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.9 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 14.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 10.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 9.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.8 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 1.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.3 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.7 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 13.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.7 | 5.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.6 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.6 | 8.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.5 | 1.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.5 | 4.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 7.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.5 | 7.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 10.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.4 | 8.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.4 | 6.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 3.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 3.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 5.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 14.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 2.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 2.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 5.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 14.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 7.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 7.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 2.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 8.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 3.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 7.0 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 17.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 2.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 5.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 5.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 2.1 | 12.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.3 | 3.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.1 | 3.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.0 | 3.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.0 | 7.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.8 | 5.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.7 | 2.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.7 | 3.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.7 | 8.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.7 | 4.7 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.6 | 12.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.5 | 4.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 7.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 6.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.4 | 0.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.4 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 5.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.4 | 2.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.4 | 2.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 12.1 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.4 | 8.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.4 | 3.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.4 | 1.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 1.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 6.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.3 | 2.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 4.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 2.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 5.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 0.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 7.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 2.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 18.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 1.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 1.5 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.2 | 0.7 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 1.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 1.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 3.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 2.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.6 | REACTOME NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Neurotransmitter Release Cycle |
| 0.2 | 1.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 3.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 2.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 5.7 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 7.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 3.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |