Project
DANIO-CODE
Navigation
Downloads
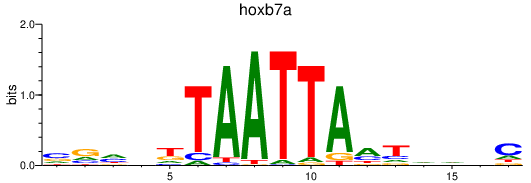
Results for hoxb7a
Z-value: 0.69
Transcription factors associated with hoxb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb7a
|
ENSDARG00000056030 | homeobox B7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb7a | dr10_dc_chr3_+_23561502_23561521 | -0.85 | 3.3e-05 | Click! |
Activity profile of hoxb7a motif
Sorted Z-values of hoxb7a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb7a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_2373601 | 2.06 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr9_+_21982644 | 1.45 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr10_-_21404605 | 1.45 |
ENSDART00000125167
|
avd
|
avidin |
| chr3_-_39912816 | 1.37 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr21_+_34053739 | 1.32 |
ENSDART00000147519
|
mtmr1b
|
myotubularin related protein 1b |
| chr16_-_43061368 | 1.31 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr3_-_19890717 | 1.26 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr18_-_43890836 | 1.23 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr21_-_13026036 | 1.21 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr1_+_22160932 | 1.20 |
ENSDART00000016488
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr6_+_9663119 | 1.20 |
ENSDART00000169544
ENSDART00000005573 |
tmem237b
|
transmembrane protein 237b |
| chr25_-_13394261 | 1.19 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr17_+_16082472 | 1.16 |
ENSDART00000133154
|
znf395a
|
zinc finger protein 395a |
| chr2_+_15379961 | 1.16 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr2_+_15379717 | 1.06 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr9_+_21982679 | 1.05 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr10_-_31129270 | 1.04 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr10_-_31129217 | 1.01 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr21_-_33998148 | 0.98 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr8_+_23806179 | 0.98 |
ENSDART00000040362
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr5_+_6005049 | 0.89 |
|
|
|
| chr7_+_7463565 | 0.88 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr21_-_33998091 | 0.86 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr2_+_15380054 | 0.86 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr21_+_34053590 | 0.81 |
ENSDART00000147519
ENSDART00000158115 ENSDART00000029599 ENSDART00000145123 |
mtmr1b
|
myotubularin related protein 1b |
| chr25_-_28630138 | 0.76 |
|
|
|
| chr4_-_765957 | 0.75 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr9_+_21982756 | 0.72 |
ENSDART00000059652
|
rev1
|
REV1, polymerase (DNA directed) |
| chr6_+_59580554 | 0.68 |
|
|
|
| chr21_-_33998117 | 0.67 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr14_+_47368951 | 0.67 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr21_-_13026077 | 0.66 |
ENSDART00000024616
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr23_-_25853364 | 0.65 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr21_-_33998051 | 0.60 |
ENSDART00000047515
ENSDART00000138575 ENSDART00000170749 |
rnf145b
|
ring finger protein 145b |
| chr18_+_14651021 | 0.58 |
ENSDART00000166679
|
cirh1a
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr17_+_16082983 | 0.57 |
ENSDART00000133154
|
znf395a
|
zinc finger protein 395a |
| chr5_+_20640396 | 0.56 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr15_-_43402935 | 0.52 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr23_+_39569262 | 0.51 |
ENSDART00000031616
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr3_-_36460498 | 0.47 |
ENSDART00000161501
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr24_-_17261929 | 0.44 |
ENSDART00000153858
|
BX005392.2
|
ENSDARG00000096996 |
| chr7_+_21006509 | 0.43 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr4_-_1952230 | 0.41 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr9_+_55395276 | 0.40 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr23_+_28882409 | 0.39 |
ENSDART00000078171
|
pex14
|
peroxisomal biogenesis factor 14 |
| chr17_-_15221715 | 0.38 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr6_+_7257043 | 0.37 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr5_-_23171454 | 0.34 |
ENSDART00000135153
|
TBC1D8B
|
TBC1 domain family member 8B |
| chr3_+_18621758 | 0.34 |
ENSDART00000156747
|
AL772362.1
|
ENSDARG00000097552 |
| KN150706v1_+_7627 | 0.33 |
|
|
|
| chr25_-_35094522 | 0.32 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr15_+_15920149 | 0.29 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr13_-_36672606 | 0.28 |
ENSDART00000179242
|
sav1
|
salvador family WW domain containing protein 1 |
| chr22_-_19077300 | 0.26 |
ENSDART00000166295
|
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr23_-_25853331 | 0.26 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr16_+_42763933 | 0.24 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr5_+_20640468 | 0.23 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr15_-_36490729 | 0.22 |
|
|
|
| chr17_-_49355906 | 0.19 |
ENSDART00000162563
|
znf292a
|
zinc finger protein 292a |
| chr6_+_59580339 | 0.19 |
|
|
|
| chr8_-_25015215 | 0.18 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr23_-_25853220 | 0.17 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr16_-_52961072 | 0.16 |
|
|
|
| chr5_+_57055662 | 0.12 |
ENSDART00000166545
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr17_+_8642686 | 0.11 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr4_-_766308 | 0.11 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr2_+_50873843 | 0.09 |
|
|
|
| chr4_-_1952201 | 0.06 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr16_+_42763696 | 0.02 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr7_+_6814762 | 0.01 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.5 | 3.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 1.0 | GO:1901741 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.4 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 2.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.7 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.4 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.8 | GO:1900746 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 0.5 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 2.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 2.9 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 3.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.5 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.2 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0017125 | deoxycytidyl transferase activity(GO:0017125) |
| 0.4 | 1.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.3 | 1.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 2.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 1.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.5 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.0 | 3.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 3.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 3.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |