Project
DANIO-CODE
Navigation
Downloads
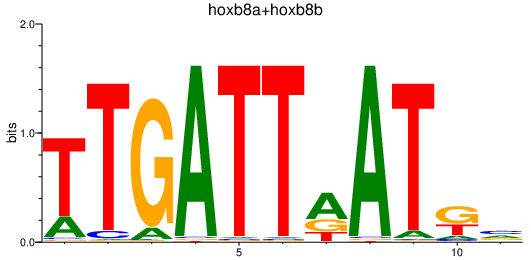
Results for hoxb8a+hoxb8b
Z-value: 1.40
Transcription factors associated with hoxb8a+hoxb8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb8b
|
ENSDARG00000054025 | homeobox B8b |
|
hoxb8a
|
ENSDARG00000056027 | homeobox B8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb8b | dr10_dc_chr12_+_27026112_27026160 | 0.89 | 4.8e-06 | Click! |
| hoxb8a | dr10_dc_chr3_+_23557320_23557417 | 0.89 | 5.2e-06 | Click! |
Activity profile of hoxb8a+hoxb8b motif
Sorted Z-values of hoxb8a+hoxb8b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb8a+hoxb8b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_5440923 | 5.00 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr13_+_30673907 | 4.60 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr3_+_30790513 | 3.65 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr20_-_29517770 | 3.53 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr7_+_42124857 | 3.47 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr25_+_24520476 | 3.40 |
|
|
|
| chr3_-_34363698 | 3.13 |
|
|
|
| chr23_+_36023748 | 3.11 |
|
|
|
| chr23_+_32573474 | 2.98 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr3_+_33168814 | 2.96 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr24_-_28814066 | 2.89 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr5_+_49103778 | 2.84 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr21_-_22510751 | 2.65 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr15_-_31687 | 2.60 |
|
|
|
| chr14_-_24463906 | 2.59 |
ENSDART00000126199
|
slit3
|
slit homolog 3 (Drosophila) |
| chr9_-_20562293 | 2.58 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr14_+_21999898 | 2.53 |
|
|
|
| chr19_+_20209561 | 2.52 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr14_+_25168063 | 2.49 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr1_+_25378105 | 2.45 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr25_+_15551288 | 2.43 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr20_-_7303735 | 2.42 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr10_-_42157835 | 2.42 |
ENSDART00000141500
ENSDART00000156626 |
BX005309.1
|
ENSDARG00000093535 |
| chr6_+_14822990 | 2.40 |
ENSDART00000149949
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr8_-_34077387 | 2.35 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr19_-_3183062 | 2.30 |
ENSDART00000122816
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr6_+_14823185 | 2.27 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr3_-_34208423 | 2.24 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr5_-_62747812 | 2.20 |
|
|
|
| chr18_+_21419735 | 2.18 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr6_-_14821305 | 2.09 |
|
|
|
| chr4_+_13811932 | 2.08 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr21_-_23271127 | 2.05 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr17_+_52736192 | 1.95 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr24_+_18804086 | 1.94 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_-_18709814 | 1.93 |
ENSDART00000145224
|
rbm47
|
RNA binding motif protein 47 |
| chr11_+_6809190 | 1.91 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr25_+_15551474 | 1.87 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr24_+_34227202 | 1.84 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr23_+_24711233 | 1.82 |
|
|
|
| chr20_-_7303804 | 1.82 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr15_+_21231406 | 1.81 |
ENSDART00000038499
|
CR626907.1
|
ENSDARG00000077872 |
| chr14_+_4489377 | 1.80 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr9_-_23406315 | 1.79 |
ENSDART00000159256
|
kif5c
|
kinesin family member 5C |
| chr11_+_3940085 | 1.79 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr16_+_21121428 | 1.66 |
|
|
|
| chr11_+_13470883 | 1.64 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr15_+_24628386 | 1.63 |
ENSDART00000148022
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr16_+_46435014 | 1.59 |
ENSDART00000144000
|
rpz2
|
rapunzel 2 |
| chr17_-_52735250 | 1.59 |
|
|
|
| chr13_+_23152038 | 1.58 |
ENSDART00000171676
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr8_-_52426509 | 1.55 |
ENSDART00000015081
|
ENSDARG00000011208
|
ENSDARG00000011208 |
| chr5_-_24365811 | 1.54 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr1_-_25377787 | 1.52 |
|
|
|
| chr22_+_18364282 | 1.50 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr5_-_35143549 | 1.50 |
ENSDART00000157881
|
ophn1
|
oligophrenin 1 |
| chr13_-_5440751 | 1.49 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr10_+_31358236 | 1.48 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr16_+_10885785 | 1.48 |
ENSDART00000161969
|
atp1a3b
|
ATPase, Na+/K+ transporting, alpha 3b polypeptide |
| chr5_+_44407763 | 1.46 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr7_+_31608828 | 1.44 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr12_-_26314881 | 1.43 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr13_-_5440558 | 1.42 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr5_-_24365560 | 1.42 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr5_+_296856 | 1.42 |
|
|
|
| chr8_+_18552850 | 1.40 |
ENSDART00000177476
|
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr2_-_57133471 | 1.38 |
|
|
|
| chr23_-_32378111 | 1.32 |
ENSDART00000143772
|
dgkaa
|
diacylglycerol kinase, alpha a |
| chr2_-_9848848 | 1.32 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr18_+_5707066 | 1.32 |
|
|
|
| chr7_-_25426346 | 1.31 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr20_+_20772543 | 1.28 |
ENSDART00000152481
|
rtn1b
|
reticulon 1b |
| chr21_+_43674754 | 1.28 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr1_+_50547214 | 1.25 |
ENSDART00000132244
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr3_+_59491893 | 1.24 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr2_-_36058327 | 1.24 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr2_-_50638153 | 1.23 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein-like 2b |
| chr16_-_50139709 | 1.22 |
|
|
|
| chr5_-_62747914 | 1.21 |
|
|
|
| chr21_+_13063614 | 1.21 |
|
|
|
| chr21_-_36886505 | 1.18 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr7_+_53485273 | 1.16 |
ENSDART00000158160
ENSDART00000163261 |
neo1a
|
neogenin 1a |
| chr1_-_46292847 | 1.13 |
ENSDART00000125032
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr7_-_18346474 | 1.12 |
|
|
|
| chr15_+_24628435 | 1.11 |
ENSDART00000139885
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr25_-_31291943 | 1.11 |
ENSDART00000175104
|
lamb4
|
laminin, beta 4 |
| chr24_-_14447773 | 1.10 |
|
|
|
| chr14_+_6647838 | 1.09 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr9_+_32267615 | 1.08 |
|
|
|
| chr3_+_23621843 | 1.08 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr19_+_5562107 | 1.07 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr12_-_34621359 | 1.07 |
|
|
|
| chr3_+_27615311 | 1.05 |
|
|
|
| chr7_-_34854886 | 1.05 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr12_-_7902815 | 1.02 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr10_-_27780050 | 1.02 |
ENSDART00000138149
|
si:dkey-33o22.1
|
si:dkey-33o22.1 |
| chr22_+_35113233 | 0.97 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr22_-_22337268 | 0.96 |
|
|
|
| chr11_-_25222937 | 0.96 |
|
|
|
| chr5_-_24365703 | 0.93 |
ENSDART00000112287
|
gas2l1
|
growth arrest-specific 2 like 1 |
| chr7_+_37105966 | 0.92 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr2_-_24947660 | 0.91 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr15_+_28269289 | 0.91 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr1_+_23093475 | 0.90 |
|
|
|
| chr22_+_35090105 | 0.89 |
|
|
|
| chr16_+_17481157 | 0.88 |
ENSDART00000173448
|
fam131bb
|
family with sequence similarity 131, member Bb |
| chr17_-_19606453 | 0.87 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr23_-_21607811 | 0.83 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr11_-_11689402 | 0.82 |
|
|
|
| chr20_-_2936289 | 0.81 |
ENSDART00000104667
|
cdk19
|
cyclin-dependent kinase 19 |
| chr3_-_16077831 | 0.79 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr21_+_8249235 | 0.79 |
ENSDART00000129749
|
psmb7
|
proteasome subunit beta 7 |
| chr3_+_23608395 | 0.77 |
|
|
|
| chr3_+_26015378 | 0.76 |
|
|
|
| chr16_+_24058139 | 0.72 |
|
|
|
| chr8_-_14446883 | 0.71 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr23_-_20412221 | 0.70 |
ENSDART00000132769
|
CR749762.2
|
ENSDARG00000093223 |
| chr17_+_45892751 | 0.69 |
|
|
|
| chr24_-_28813986 | 0.67 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr6_-_23830912 | 0.66 |
|
|
|
| chr24_-_25021505 | 0.64 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr3_+_23621561 | 0.62 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr15_+_8791540 | 0.62 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr24_+_24316486 | 0.62 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr7_-_32977618 | 0.60 |
ENSDART00000052389
|
itga11a
|
integrin, alpha 11a |
| chr9_+_32267297 | 0.57 |
|
|
|
| chr5_+_19737802 | 0.53 |
ENSDART00000153643
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr8_-_18546957 | 0.53 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr3_+_7727322 | 0.52 |
ENSDART00000160328
|
zgc:109949
|
zgc:109949 |
| chr8_+_40604188 | 0.51 |
|
|
|
| chr14_+_35890811 | 0.50 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr12_+_39496010 | 0.49 |
|
|
|
| chr10_+_26609369 | 0.48 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr17_+_23102259 | 0.44 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr13_+_23152160 | 0.44 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr20_+_20772488 | 0.43 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr2_+_31682629 | 0.43 |
ENSDART00000143914
|
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr11_+_23012136 | 0.40 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr7_+_21006803 | 0.40 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr21_-_27301938 | 0.39 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr6_+_29096734 | 0.36 |
|
|
|
| chr10_+_36082726 | 0.35 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr13_+_8272812 | 0.34 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr20_-_7303680 | 0.33 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr24_-_28813917 | 0.33 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr5_+_36332564 | 0.33 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr24_-_32115473 | 0.32 |
ENSDART00000168419
|
rsu1
|
Ras suppressor protein 1 |
| chr8_-_18547147 | 0.32 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr24_-_14447136 | 0.32 |
|
|
|
| chr10_+_31358307 | 0.32 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr25_+_32419062 | 0.31 |
|
|
|
| chr24_+_24316408 | 0.29 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_-_22466834 | 0.29 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr4_+_13430131 | 0.29 |
ENSDART00000137200
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr2_+_1881022 | 0.28 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr11_-_3940007 | 0.28 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr21_-_26446246 | 0.24 |
ENSDART00000112410
|
ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr12_-_11419332 | 0.24 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr25_+_19889409 | 0.23 |
ENSDART00000178266
ENSDART00000121226 |
FO704882.1
|
ENSDARG00000085826 |
| chr7_+_51879116 | 0.23 |
ENSDART00000098712
|
TMEM208
|
transmembrane protein 208 |
| chr13_-_36495730 | 0.22 |
ENSDART00000165629
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr24_+_24316346 | 0.20 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_-_686007 | 0.16 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr8_-_18547116 | 0.16 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr24_-_28814284 | 0.14 |
ENSDART00000003503
|
col11a1a
|
collagen, type XI, alpha 1a |
| chr23_-_6793566 | 0.12 |
ENSDART00000171714
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr7_+_38540519 | 0.10 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| KN149949v1_+_25867 | 0.08 |
|
|
|
| KN149949v1_+_25953 | 0.08 |
|
|
|
| chr23_-_31033917 | 0.06 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr10_-_33212897 | 0.06 |
ENSDART00000081170
|
cux1a
|
cut-like homeobox 1a |
| chr3_+_35277555 | 0.06 |
|
|
|
| chr19_+_12525407 | 0.05 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr14_+_7626683 | 0.04 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr11_-_27832530 | 0.04 |
ENSDART00000172937
|
BX248318.1
|
ENSDARG00000104418 |
| chr11_+_13470759 | 0.01 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.9 | 2.6 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.6 | 5.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 2.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.5 | 7.9 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.5 | 1.9 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.4 | 1.3 | GO:0002280 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.4 | 1.2 | GO:0032475 | otolith formation(GO:0032475) |
| 0.4 | 1.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 1.8 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.3 | 1.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.3 | 1.9 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 0.3 | 2.2 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.3 | 1.8 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.3 | 1.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.3 | 1.5 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.2 | 1.8 | GO:0060827 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.2 | 3.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 3.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.6 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 1.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 2.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.7 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.6 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.5 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 3.6 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 2.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.9 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 7.2 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 5.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.9 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 4.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 3.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.6 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 4.6 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.4 | 1.4 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 2.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 3.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.5 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 9.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.1 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 8.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 15.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 3.0 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 3.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 2.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.6 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |