Project
DANIO-CODE
Navigation
Downloads
Results for hoxb9a
Z-value: 0.43
Transcription factors associated with hoxb9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb9a
|
ENSDARG00000056023 | homeobox B9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb9a | dr10_dc_chr3_+_23546802_23546908 | -0.30 | 2.6e-01 | Click! |
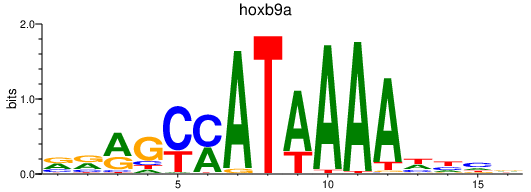
Activity profile of hoxb9a motif
Sorted Z-values of hoxb9a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb9a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_44853207 | 0.75 |
ENSDART00000169466
|
scarb1
|
scavenger receptor class B, member 1 |
| chr6_+_49552893 | 0.71 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr2_-_45810787 | 0.69 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr16_-_17678748 | 0.63 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr13_-_15662476 | 0.62 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr1_-_8801877 | 0.60 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr3_-_29779725 | 0.59 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr6_+_23476669 | 0.58 |
|
|
|
| chr4_+_12293403 | 0.54 |
ENSDART00000061070
|
mkrn1
|
makorin, ring finger protein, 1 |
| chr15_+_14656797 | 0.53 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr5_-_31257158 | 0.49 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr5_-_23211957 | 0.48 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr3_-_29779598 | 0.47 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr13_-_15662599 | 0.45 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr8_+_3373066 | 0.44 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr20_+_27188382 | 0.43 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr14_+_24543399 | 0.43 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr23_+_22858063 | 0.43 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr2_-_45810868 | 0.40 |
ENSDART00000135665
ENSDART00000075080 |
prpf38b
|
pre-mRNA processing factor 38B |
| chr22_+_2735606 | 0.40 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr21_-_3548863 | 0.39 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr5_-_57053687 | 0.39 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr21_-_11877525 | 0.38 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr2_+_23383492 | 0.37 |
ENSDART00000170669
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr4_+_16020464 | 0.37 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr8_+_3372903 | 0.37 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr21_-_38979792 | 0.36 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr24_-_38758093 | 0.36 |
|
|
|
| chr22_+_15317622 | 0.36 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr8_-_1601007 | 0.33 |
ENSDART00000159705
|
dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr18_+_35198798 | 0.31 |
ENSDART00000098292
|
cfap45
|
cilia and flagella associated protein 45 |
| chr14_+_14536088 | 0.29 |
ENSDART00000158291
|
slbp
|
stem-loop binding protein |
| chr11_+_24076576 | 0.27 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr1_-_8565788 | 0.26 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr24_-_32110851 | 0.26 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr15_+_44069851 | 0.26 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr20_-_31593936 | 0.23 |
ENSDART00000153193
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr10_-_44441197 | 0.21 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr5_+_9925967 | 0.21 |
ENSDART00000174914
|
BX511089.4
|
ENSDARG00000106012 |
| chr25_+_22474794 | 0.21 |
ENSDART00000152096
|
stra6
|
stimulated by retinoic acid 6 |
| chr5_-_31257031 | 0.20 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr3_-_29779552 | 0.20 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr20_-_31594412 | 0.20 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr15_+_14656886 | 0.20 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr18_+_8362131 | 0.20 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| KN149795v1_-_10712 | 0.19 |
|
|
|
| chr20_-_34894930 | 0.18 |
|
|
|
| chr20_-_31593772 | 0.18 |
ENSDART00000153437
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr5_-_53777415 | 0.18 |
ENSDART00000169270
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr6_+_12849673 | 0.18 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr2_+_23383380 | 0.17 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr17_-_49980590 | 0.16 |
ENSDART00000106562
|
zgc:113886
|
zgc:113886 |
| chr10_+_16111920 | 0.16 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr23_-_35904414 | 0.14 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr20_-_31594202 | 0.14 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr1_+_25411387 | 0.14 |
ENSDART00000127154
|
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr6_+_12849629 | 0.14 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr24_+_31396974 | 0.14 |
ENSDART00000168837
|
fam168b
|
family with sequence similarity 168, member B |
| chr8_+_3373146 | 0.14 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr6_+_4379989 | 0.13 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr2_+_23383257 | 0.12 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr21_-_1339525 | 0.12 |
|
|
|
| chr6_+_30385400 | 0.12 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr23_-_36023579 | 0.12 |
|
|
|
| chr6_+_30385355 | 0.12 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr1_-_8801818 | 0.12 |
ENSDART00000008682
ENSDART00000157814 |
micall2b
|
mical-like 2b |
| chr11_+_44002664 | 0.12 |
|
|
|
| chr5_+_23583233 | 0.11 |
ENSDART00000177458
|
tp53
|
tumor protein p53 |
| chr12_+_26931418 | 0.11 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr23_-_36023622 | 0.11 |
|
|
|
| chr10_-_44441390 | 0.11 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr20_+_20887959 | 0.11 |
|
|
|
| chr20_-_34767188 | 0.10 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr10_-_11011134 | 0.10 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr23_+_22857958 | 0.10 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr8_-_1601118 | 0.10 |
ENSDART00000026747
|
dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr14_-_30642610 | 0.10 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr24_+_17044956 | 0.10 |
|
|
|
| chr13_+_15802079 | 0.09 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr18_-_17085956 | 0.09 |
ENSDART00000042496
|
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr15_-_26998580 | 0.09 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| KN150442v1_-_37820 | 0.09 |
ENSDART00000158371
|
ptmab
|
prothymosin, alpha b |
| chr12_+_19183576 | 0.08 |
ENSDART00000066391
|
csnk1e
|
casein kinase 1, epsilon |
| chr3_+_36504142 | 0.08 |
ENSDART00000170013
|
gspt1l
|
G1 to S phase transition 1, like |
| chr20_-_2696401 | 0.08 |
ENSDART00000104606
|
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr9_+_33247126 | 0.08 |
|
|
|
| chr6_-_3821922 | 0.08 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr24_+_31751950 | 0.07 |
ENSDART00000156493
|
CT737162.1
|
ENSDARG00000096998 |
| chr3_-_25142506 | 0.07 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr11_-_43409685 | 0.07 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr14_-_30642549 | 0.07 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr24_+_29278310 | 0.07 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr23_+_44937093 | 0.07 |
|
|
|
| chr5_-_53776769 | 0.06 |
ENSDART00000164595
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr13_+_36459562 | 0.06 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr14_+_5554581 | 0.06 |
ENSDART00000041279
|
tubb4b
|
tubulin, beta 4B class IVb |
| chr3_+_23604393 | 0.06 |
|
|
|
| chr23_-_45265411 | 0.06 |
|
|
|
| chr5_+_23582805 | 0.05 |
ENSDART00000137298
|
tp53
|
tumor protein p53 |
| chr19_+_20174462 | 0.05 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr24_-_32110808 | 0.04 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr25_+_15983777 | 0.04 |
ENSDART00000165598
ENSDART00000061753 |
far1
|
fatty acyl CoA reductase 1 |
| chr3_-_25142301 | 0.04 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr3_-_23558171 | 0.04 |
|
|
|
| chr1_+_44971899 | 0.03 |
ENSDART00000149565
|
trappc5
|
trafficking protein particle complex 5 |
| chr2_-_6545431 | 0.03 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr1_+_29027527 | 0.03 |
ENSDART00000142184
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr3_-_25142556 | 0.03 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr16_-_28333633 | 0.03 |
ENSDART00000121671
|
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr20_-_2624028 | 0.02 |
ENSDART00000145335
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr1_+_51846516 | 0.02 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr5_+_71409912 | 0.02 |
ENSDART00000165835
|
ddx54
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 |
| chr23_+_39713307 | 0.02 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr7_-_32743153 | 0.02 |
|
|
|
| chr22_+_15317535 | 0.02 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr25_+_19992328 | 0.02 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr4_+_14107347 | 0.02 |
ENSDART00000150748
|
CU469420.1
|
ENSDARG00000086259 |
| chr5_+_19506512 | 0.01 |
ENSDART00000047841
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr19_-_27864288 | 0.01 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| KN150196v1_+_15294 | 0.01 |
|
|
|
| chr1_-_37639802 | 0.01 |
|
|
|
| chr22_-_3954917 | 0.01 |
ENSDART00000162523
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr17_-_21037758 | 0.01 |
ENSDART00000048853
|
ube2d1a
|
ubiquitin-conjugating enzyme E2D 1a |
| chr17_-_36894361 | 0.01 |
ENSDART00000176989
|
myo6b
|
myosin VIb |
| chr5_-_53777106 | 0.00 |
ENSDART00000164067
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr23_+_13368050 | 0.00 |
ENSDART00000139475
|
samd10b
|
sterile alpha motif domain containing 10b |
| chr4_+_295110 | 0.00 |
ENSDART00000067489
|
si:ch211-51e12.7
|
si:ch211-51e12.7 |
| chr22_-_20963723 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.9 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.7 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.1 | 0.3 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.2 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 1.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |