Project
DANIO-CODE
Navigation
Downloads
Results for hoxc11b
Z-value: 1.03
Transcription factors associated with hoxc11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11b
|
ENSDARG00000102631 | homeobox C11b |
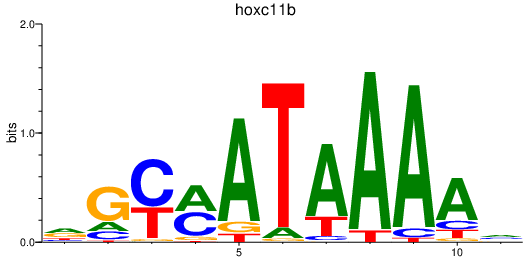
Activity profile of hoxc11b motif
Sorted Z-values of hoxc11b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45950530 | 4.41 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr3_+_26896869 | 4.27 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr23_+_10211543 | 3.95 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr23_+_39713307 | 3.57 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr16_-_39620777 | 3.21 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr18_-_15404998 | 3.17 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr24_+_5205878 | 3.15 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr18_+_22617072 | 3.15 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr14_-_24463906 | 2.80 |
ENSDART00000126199
|
slit3
|
slit homolog 3 (Drosophila) |
| chr23_+_25428372 | 2.67 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr9_-_48673183 | 2.62 |
ENSDART00000140185
ENSDART00000134185 |
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr5_-_39910235 | 2.57 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr6_+_56157608 | 2.44 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr7_+_35769973 | 2.41 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| chr2_+_20748431 | 2.40 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr8_-_44305494 | 2.38 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr18_+_26437604 | 2.36 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr8_-_38126693 | 2.35 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr18_-_15405161 | 2.34 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr23_+_36023748 | 2.34 |
|
|
|
| chr18_+_730277 | 2.33 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr14_+_4169846 | 2.31 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr21_+_25199691 | 2.28 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr9_+_17298410 | 2.26 |
ENSDART00000048548
|
scel
|
sciellin |
| chr2_-_30675594 | 2.24 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr10_-_10372266 | 2.19 |
|
|
|
| chr7_-_26035308 | 2.19 |
ENSDART00000131906
|
zgc:77439
|
zgc:77439 |
| chr6_-_39315024 | 2.15 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr19_+_22478256 | 2.10 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr23_+_22977031 | 2.01 |
|
|
|
| chr15_-_25592228 | 1.79 |
ENSDART00000157498
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr6_+_35378839 | 1.79 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr1_+_45503061 | 1.76 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr3_+_61924544 | 1.75 |
ENSDART00000090370
|
noxo1a
|
NADPH oxidase organizer 1a |
| chr20_+_51385187 | 1.74 |
|
|
|
| chr2_+_53253623 | 1.73 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr9_-_1954910 | 1.73 |
ENSDART00000140438
|
BX322661.1
|
ENSDARG00000094186 |
| chr25_-_13518274 | 1.70 |
ENSDART00000165510
|
fa2h
|
fatty acid 2-hydroxylase |
| chr25_+_19992389 | 1.69 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr15_-_1858350 | 1.67 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr17_-_52735250 | 1.67 |
|
|
|
| chr8_+_47644421 | 1.66 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr18_+_45893199 | 1.63 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr5_-_40707316 | 1.63 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr3_-_19050721 | 1.63 |
ENSDART00000131503
|
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr3_+_23563620 | 1.63 |
ENSDART00000147022
|
hoxb7a
|
homeobox B7a |
| chr17_-_23707863 | 1.62 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr3_+_23580269 | 1.62 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr17_-_2513630 | 1.58 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr16_+_21436660 | 1.55 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr4_+_5733160 | 1.51 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr16_-_33105847 | 1.49 |
|
|
|
| chr10_-_10906027 | 1.47 |
ENSDART00000122657
|
nrarpa
|
notch-regulated ankyrin repeat protein a |
| chr21_-_17259886 | 1.46 |
ENSDART00000114877
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr23_+_7445760 | 1.45 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr16_-_55320569 | 1.44 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr14_-_31128891 | 1.44 |
|
|
|
| chr17_+_52736192 | 1.42 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr23_+_27141681 | 1.39 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr5_-_41672394 | 1.39 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr10_-_22834248 | 1.32 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr24_-_16884946 | 1.31 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr21_-_3548719 | 1.30 |
ENSDART00000137844
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr2_-_40170303 | 1.29 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr14_+_34626233 | 1.27 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr19_-_19290260 | 1.26 |
|
|
|
| chr20_+_26981663 | 1.24 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr5_+_69383382 | 1.21 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| KN150230v1_-_36595 | 1.20 |
|
|
|
| chr6_-_50705420 | 1.19 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr11_+_42265857 | 1.17 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr14_+_45895103 | 1.17 |
|
|
|
| chr12_-_27032151 | 1.17 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr1_+_27174549 | 1.16 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr17_-_37040941 | 1.16 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr23_-_7740845 | 1.15 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr20_+_52740555 | 1.14 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr25_+_30701751 | 1.13 |
|
|
|
| chr6_-_60152693 | 1.10 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr21_+_10663517 | 1.09 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr4_-_13394059 | 1.07 |
ENSDART00000175909
ENSDART00000174861 |
BX511038.1
|
ENSDARG00000108089 |
| chr24_+_30386569 | 1.07 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr22_-_22696091 | 1.07 |
|
|
|
| chr13_+_12257610 | 1.07 |
ENSDART00000086525
|
atp10d
|
ATPase, class V, type 10D |
| chr17_+_52736535 | 1.07 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr16_-_21238288 | 1.07 |
ENSDART00000139737
|
cbx3b
|
chromobox homolog 3b |
| chr17_-_52735615 | 1.05 |
|
|
|
| chr13_+_4276783 | 1.04 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr11_-_18166056 | 1.03 |
ENSDART00000155752
|
sfmbt1
|
Scm-like with four mbt domains 1 |
| chr23_+_22858531 | 1.02 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr7_-_23474701 | 1.02 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr5_-_66293492 | 1.01 |
|
|
|
| chr22_-_10540794 | 0.99 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr25_-_22821132 | 0.98 |
|
|
|
| chr9_-_23346038 | 0.98 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr12_-_18839556 | 0.96 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr23_+_24711233 | 0.95 |
|
|
|
| chr6_-_60152594 | 0.95 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr10_-_31619761 | 0.94 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr23_+_35954132 | 0.91 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr10_-_42157835 | 0.90 |
ENSDART00000141500
ENSDART00000156626 |
BX005309.1
|
ENSDARG00000093535 |
| chr5_+_64411584 | 0.88 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr5_+_23741791 | 0.87 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr4_+_27141061 | 0.85 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr17_+_24428093 | 0.84 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr19_+_20189224 | 0.83 |
ENSDART00000163611
|
hoxa4a
|
homeobox A4a |
| chr19_-_27152063 | 0.83 |
ENSDART00000109258
|
csnk2b
|
casein kinase 2, beta polypeptide |
| chr23_-_10906094 | 0.82 |
|
|
|
| chr19_+_42899678 | 0.81 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr4_+_119964 | 0.81 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr12_+_10096495 | 0.79 |
ENSDART00000152212
ENSDART00000127343 |
smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr25_+_5567388 | 0.79 |
|
|
|
| chr17_+_15780370 | 0.78 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr17_+_52736844 | 0.78 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr5_+_19802818 | 0.76 |
|
|
|
| chr10_+_2559774 | 0.76 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr25_+_34791665 | 0.75 |
|
|
|
| chr8_-_18868986 | 0.75 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr8_+_25984588 | 0.75 |
|
|
|
| chr21_+_26695270 | 0.74 |
|
|
|
| chr5_+_52934865 | 0.74 |
ENSDART00000161682
|
BX323994.2
|
ENSDARG00000100423 |
| chr13_-_24779651 | 0.73 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr19_+_20174462 | 0.71 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr19_-_36146250 | 0.71 |
ENSDART00000066712
|
trappc3
|
trafficking protein particle complex 3 |
| chr6_+_30716955 | 0.70 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr5_-_70688185 | 0.67 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr18_+_26437724 | 0.67 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr4_+_5246465 | 0.67 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr11_+_36781811 | 0.67 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr8_-_44305846 | 0.67 |
ENSDART00000144497
|
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr6_+_4379989 | 0.66 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr10_-_30016761 | 0.66 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr1_+_966385 | 0.65 |
ENSDART00000051919
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr24_-_8592102 | 0.64 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr25_-_34468195 | 0.63 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr20_+_20887959 | 0.63 |
|
|
|
| chr23_+_11734349 | 0.63 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr22_-_26307952 | 0.61 |
|
|
|
| chr24_-_6129575 | 0.60 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr7_+_60992745 | 0.60 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr15_-_25457193 | 0.59 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr4_-_5643440 | 0.59 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr11_+_18020191 | 0.59 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr11_-_24190842 | 0.57 |
ENSDART00000103752
|
dvl1b
|
dishevelled segment polarity protein 1b |
| chr7_-_18256512 | 0.55 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr7_+_31608878 | 0.55 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr13_-_35908743 | 0.55 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr24_-_38758093 | 0.54 |
|
|
|
| chr17_-_18869111 | 0.54 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr5_-_8568715 | 0.53 |
ENSDART00000099891
|
atp5ib
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Eb |
| chr3_+_25023544 | 0.53 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr12_-_24974450 | 0.53 |
ENSDART00000163775
ENSDART00000159167 |
socs5a
|
suppressor of cytokine signaling 5a |
| chr23_+_4544178 | 0.52 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr11_-_18305730 | 0.51 |
ENSDART00000155474
|
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| chr10_-_24401876 | 0.51 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr1_+_32669248 | 0.50 |
ENSDART00000133870
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr20_-_16553010 | 0.50 |
|
|
|
| chr24_-_8592157 | 0.49 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr3_+_24003840 | 0.49 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr2_+_23600656 | 0.49 |
|
|
|
| chr5_+_58017820 | 0.47 |
ENSDART00000083015
|
ccdc84
|
coiled-coil domain containing 84 |
| KN150183v1_+_12947 | 0.46 |
|
|
|
| chr20_-_51454257 | 0.45 |
ENSDART00000023064
|
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr20_-_34767302 | 0.45 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr10_+_44853121 | 0.44 |
ENSDART00000167952
|
scarb1
|
scavenger receptor class B, member 1 |
| chr25_+_15983777 | 0.44 |
ENSDART00000165598
ENSDART00000061753 |
far1
|
fatty acyl CoA reductase 1 |
| chr18_-_15361133 | 0.44 |
ENSDART00000019818
|
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr14_+_4169371 | 0.43 |
ENSDART00000136665
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr17_-_9794215 | 0.40 |
|
|
|
| chr17_+_21270766 | 0.39 |
|
|
|
| chr3_+_18263447 | 0.38 |
ENSDART00000127796
|
gaa
|
glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) |
| chr9_+_33049445 | 0.38 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr13_+_36459706 | 0.37 |
ENSDART00000138940
|
gmfb
|
glia maturation factor, beta |
| chr1_+_51846516 | 0.36 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr23_+_44543892 | 0.35 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr9_+_33049549 | 0.35 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr12_-_20251183 | 0.35 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr12_+_26930895 | 0.35 |
ENSDART00000152975
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr4_+_14658179 | 0.34 |
ENSDART00000168152
|
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_-_51658383 | 0.34 |
ENSDART00000132638
|
ENSDARG00000059948
|
ENSDARG00000059948 |
| chr3_-_39117801 | 0.33 |
ENSDART00000102678
|
nmt1a
|
N-myristoyltransferase 1a |
| chr23_+_22858650 | 0.32 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr19_-_15324958 | 0.32 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr20_-_34767188 | 0.32 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr12_-_10372346 | 0.31 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr7_+_35770134 | 0.30 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| chr16_+_13993746 | 0.29 |
ENSDART00000101304
|
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr24_-_21366493 | 0.29 |
ENSDART00000109587
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr6_-_12060971 | 0.28 |
ENSDART00000155685
|
pkp4
|
plakophilin 4 |
| chr7_+_30874387 | 0.28 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr25_+_7359375 | 0.28 |
|
|
|
| chr22_-_11618612 | 0.28 |
ENSDART00000109596
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr2_+_1881022 | 0.27 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr1_+_33510660 | 0.27 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr6_+_60152913 | 0.26 |
|
|
|
| chr10_-_25629613 | 0.26 |
ENSDART00000131640
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr3_+_27655753 | 0.25 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr3_+_24059652 | 0.23 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr10_-_42155790 | 0.23 |
|
|
|
| chr14_+_45895037 | 0.22 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 1.0 | 3.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.6 | 3.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.6 | 2.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.5 | 4.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.5 | 2.7 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.4 | 2.0 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 2.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.4 | 1.4 | GO:0048618 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.4 | 1.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 2.2 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 1.1 | GO:0006210 | thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.2 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 0.7 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 1.2 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.2 | 1.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.2 | 1.4 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 4.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.2 | 2.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 1.3 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 0.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 0.6 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.2 | 0.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 0.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 1.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 2.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 3.6 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 0.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 1.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 2.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.6 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.6 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.8 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 4.5 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 0.7 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 2.4 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.3 | GO:1900619 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.2 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.9 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 3.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.6 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 3.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.4 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.3 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 2.3 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 2.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 2.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 5.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 6.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 9.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.2 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.7 | 2.0 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.5 | 1.6 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.5 | 2.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 2.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.3 | 1.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 2.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 2.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 1.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 4.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 3.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 0.6 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.2 | 1.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 2.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 1.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 4.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.8 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 2.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 5.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 3.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 15.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.8 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 9.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 3.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 3.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |