Project
DANIO-CODE
Navigation
Downloads
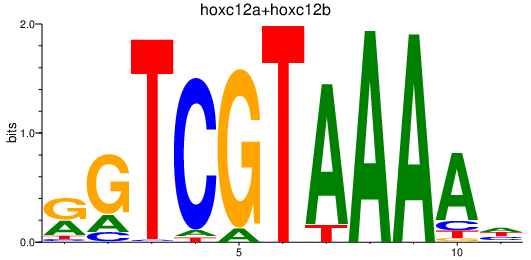
Results for hoxc12a+hoxc12b
Z-value: 0.95
Transcription factors associated with hoxc12a+hoxc12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc12a
|
ENSDARG00000070352 | homeobox C12a |
|
hoxc12b
|
ENSDARG00000103133 | homeobox C12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc12a | dr10_dc_chr23_+_35964754_35964817 | 0.67 | 4.3e-03 | Click! |
| hoxc12b | dr10_dc_chr11_-_2133416_2133453 | 0.67 | 4.4e-03 | Click! |
Activity profile of hoxc12a+hoxc12b motif
Sorted Z-values of hoxc12a+hoxc12b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc12a+hoxc12b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_35997065 | 3.52 |
ENSDART00000153910
|
BX005254.4
|
ENSDARG00000097150 |
| chr7_-_23601966 | 2.50 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr3_+_23580269 | 2.42 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr23_+_36010592 | 2.39 |
ENSDART00000137507
|
hoxc3a
|
homeo box C3a |
| chr23_+_35988395 | 2.33 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr12_+_2642541 | 2.26 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr9_-_39195468 | 2.12 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr15_-_14616083 | 2.06 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr3_-_23444371 | 1.96 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr17_-_23707863 | 1.95 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr11_-_2107054 | 1.92 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr3_-_6078015 | 1.87 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr14_-_36523075 | 1.84 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr19_+_20181908 | 1.77 |
ENSDART00000158987
|
hoxa4a
|
homeobox A4a |
| chr12_-_10001043 | 1.73 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr1_+_30215209 | 1.71 |
ENSDART00000164520
|
itga6b
|
integrin, alpha 6b |
| chr12_-_26760324 | 1.70 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr23_+_35996779 | 1.68 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr24_-_16884946 | 1.63 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr3_+_23577103 | 1.57 |
ENSDART00000025449
|
hoxb5a
|
homeobox B5a |
| chr2_-_10555152 | 1.54 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr12_+_27038210 | 1.54 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr24_+_21395671 | 1.52 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr12_+_40919018 | 1.52 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr21_+_20678383 | 1.52 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr16_+_21104310 | 1.49 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr23_+_35988426 | 1.48 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr19_+_5399813 | 1.44 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr3_-_23558171 | 1.44 |
|
|
|
| chr9_+_44233139 | 1.42 |
ENSDART00000162406
|
itga4
|
integrin alpha 4 |
| chr25_-_17906512 | 1.41 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr6_+_10630329 | 1.39 |
ENSDART00000174807
|
BX546447.1
|
ENSDARG00000108699 |
| chr23_-_28099284 | 1.38 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr4_-_13996902 | 1.35 |
ENSDART00000147955
|
prickle1b
|
prickle homolog 1b |
| chr15_-_18159904 | 1.35 |
ENSDART00000170874
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr24_-_31285981 | 1.34 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr21_+_13031158 | 1.31 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr3_-_23580055 | 1.27 |
|
|
|
| chr1_-_33880029 | 1.24 |
ENSDART00000004361
|
klf5a
|
Kruppel-like factor 5a |
| chr10_-_33678206 | 1.16 |
ENSDART00000128049
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr12_+_27038298 | 1.10 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr19_+_5399877 | 1.07 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr19_+_20189224 | 1.01 |
ENSDART00000163611
|
hoxa4a
|
homeobox A4a |
| chr11_-_35788900 | 1.00 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr20_+_48971948 | 1.00 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr7_-_18256512 | 1.00 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr14_-_27754549 | 0.99 |
ENSDART00000135337
|
zgc:64189
|
zgc:64189 |
| chr3_+_37645350 | 0.97 |
ENSDART00000134524
|
rps11
|
ribosomal protein S11 |
| chr10_+_2559774 | 0.96 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr23_-_28099391 | 0.96 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr3_+_23561215 | 0.92 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr22_-_29788268 | 0.89 |
|
|
|
| chr6_-_31160132 | 0.89 |
ENSDART00000153592
|
CR847887.1
|
ENSDARG00000097226 |
| chr8_+_3391493 | 0.88 |
ENSDART00000085993
|
pxnb
|
paxillin b |
| chr21_-_18981828 | 0.85 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr1_-_22144014 | 0.84 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr3_+_23580216 | 0.84 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr14_+_30568242 | 0.83 |
|
|
|
| chr21_+_13764828 | 0.81 |
ENSDART00000171306
|
stxbp1a
|
syntaxin binding protein 1a |
| chr7_+_20700070 | 0.80 |
|
|
|
| chr23_-_28099359 | 0.79 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr16_+_5284778 | 0.79 |
ENSDART00000156685
ENSDART00000156765 |
soga3a
soga3a
|
SOGA family member 3a SOGA family member 3a |
| chr23_+_25224499 | 0.77 |
|
|
|
| chr9_+_55778669 | 0.76 |
|
|
|
| chr3_+_59560881 | 0.71 |
ENSDART00000084738
|
ppp1r27a
|
protein phosphatase 1, regulatory subunit 27a |
| KN150230v1_-_36595 | 0.70 |
|
|
|
| chr3_-_45237479 | 0.65 |
ENSDART00000160923
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr12_-_22438379 | 0.65 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr23_+_35988366 | 0.64 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr15_+_29183251 | 0.63 |
ENSDART00000153609
|
capns1b
|
calpain, small subunit 1 b |
| chr23_-_28099235 | 0.62 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr7_+_28896401 | 0.62 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr6_+_4091384 | 0.62 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr12_-_48562929 | 0.61 |
ENSDART00000063438
|
CABZ01079984.1
|
uncharacterized protein C7orf62 homolog |
| chr2_-_55673404 | 0.60 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr3_+_23561306 | 0.60 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr14_+_35351559 | 0.58 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr6_-_28231995 | 0.56 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr17_-_51110171 | 0.55 |
ENSDART00000088185
|
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr3_+_23561502 | 0.54 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr25_-_17906739 | 0.50 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr11_+_14373731 | 0.49 |
ENSDART00000161879
|
BX571942.1
|
ENSDARG00000099220 |
| chr14_+_31682159 | 0.47 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr25_+_17906724 | 0.46 |
|
|
|
| chr6_-_53333648 | 0.45 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr8_+_48888672 | 0.44 |
|
|
|
| chr15_-_19508230 | 0.44 |
ENSDART00000114936
|
esamb
|
endothelial cell adhesion molecule b |
| chr22_+_10411257 | 0.44 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr13_+_33138144 | 0.43 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr22_-_9706237 | 0.40 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr6_+_4379989 | 0.39 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr15_-_19314712 | 0.37 |
ENSDART00000092705
|
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| KN150222v1_+_74557 | 0.36 |
|
|
|
| chr16_+_38709412 | 0.36 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr20_+_32646617 | 0.34 |
|
|
|
| chr9_-_3675527 | 0.33 |
|
|
|
| chr1_-_7417472 | 0.32 |
ENSDART00000161938
ENSDART00000163307 |
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr20_+_36098607 | 0.32 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr11_+_6648811 | 0.31 |
ENSDART00000172061
|
CR381534.1
|
ENSDARG00000098564 |
| chr16_-_6304533 | 0.31 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr2_+_24851910 | 0.31 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr2_-_6351592 | 0.30 |
|
|
|
| chr11_-_27832530 | 0.29 |
ENSDART00000172937
|
BX248318.1
|
ENSDARG00000104418 |
| chr2_+_54966412 | 0.27 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr13_-_44601523 | 0.26 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr21_+_13764917 | 0.26 |
ENSDART00000015629
|
stxbp1a
|
syntaxin binding protein 1a |
| chr2_-_39793399 | 0.25 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr19_+_20189194 | 0.25 |
ENSDART00000163611
|
hoxa4a
|
homeobox A4a |
| chr16_+_10538946 | 0.25 |
ENSDART00000173132
|
vars
|
valyl-tRNA synthetase |
| chr17_-_32913287 | 0.23 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr25_+_17906515 | 0.21 |
|
|
|
| chr3_+_24059652 | 0.20 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr1_-_45972732 | 0.20 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr2_-_37891963 | 0.19 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr9_+_20672966 | 0.19 |
ENSDART00000145111
|
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr6_+_4091251 | 0.19 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr18_-_21759607 | 0.16 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr11_+_6138968 | 0.15 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr16_-_10425382 | 0.15 |
ENSDART00000104025
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr1_-_43936717 | 0.14 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr2_-_19181629 | 0.13 |
ENSDART00000165782
|
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr19_-_45499458 | 0.13 |
|
|
|
| chr5_-_28888704 | 0.10 |
|
|
|
| chr25_+_3168376 | 0.09 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr23_+_8862155 | 0.05 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr19_-_18671902 | 0.05 |
|
|
|
| chr5_-_64637049 | 0.05 |
ENSDART00000164649
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr15_+_27454643 | 0.04 |
ENSDART00000164887
ENSDART00000018603 |
tbx4
|
T-box 4 |
| chr21_+_13030816 | 0.04 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr5_+_31889674 | 0.03 |
ENSDART00000165417
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr13_-_9543393 | 0.02 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr16_-_47491350 | 0.02 |
ENSDART00000149723
|
sept7b
|
septin 7b |
| chr23_+_36020128 | 0.02 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr7_+_27626968 | 0.02 |
ENSDART00000178912
|
tub
|
tubby bipartite transcription factor |
| chr1_-_43936611 | 0.01 |
ENSDART00000170912
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr7_+_20699940 | 0.01 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.6 | 2.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.4 | 1.7 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 1.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.6 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 2.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 3.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 2.1 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.3 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.2 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.4 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.0 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 18.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.1 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 3.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.0 | 0.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 2.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 1.2 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.8 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.3 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.1 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.6 | 1.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 31.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.3 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |