Project
DANIO-CODE
Navigation
Downloads
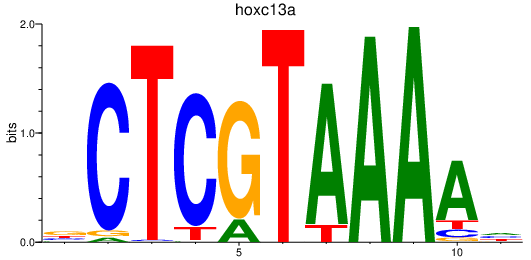
Results for hoxc13a
Z-value: 1.73
Transcription factors associated with hoxc13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc13a
|
ENSDARG00000070353 | homeobox C13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc13a | dr10_dc_chr23_+_35954132_35954180 | 0.70 | 2.3e-03 | Click! |
Activity profile of hoxc13a motif
Sorted Z-values of hoxc13a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc13a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_30234718 | 6.71 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr14_+_23917724 | 5.74 |
ENSDART00000138082
ENSDART00000079215 |
stc2a
|
stanniocalcin 2a |
| chr14_-_2008649 | 5.29 |
ENSDART00000161817
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr25_+_30699938 | 5.17 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr9_-_1964814 | 4.98 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr22_-_3661536 | 4.36 |
ENSDART00000153634
|
FP565432.1
|
ENSDARG00000097145 |
| chr7_+_23843682 | 4.17 |
ENSDART00000173899
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr9_-_1958795 | 4.01 |
ENSDART00000138730
|
hoxd3a
|
homeobox D3a |
| chr4_-_16556116 | 3.90 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_-_23601966 | 3.70 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr2_+_21067708 | 3.64 |
ENSDART00000148400
ENSDART00000021168 |
rxrga
|
retinoid x receptor, gamma a |
| chr1_-_23765358 | 3.60 |
|
|
|
| chr23_-_29138952 | 3.40 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr6_+_10630329 | 3.32 |
ENSDART00000174807
|
BX546447.1
|
ENSDARG00000108699 |
| chr14_+_36156947 | 3.23 |
|
|
|
| chr6_-_23193752 | 3.22 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr2_+_57136624 | 3.13 |
ENSDART00000159912
|
ENSDARG00000105208
|
ENSDARG00000105208 |
| chr22_+_12327849 | 3.06 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr19_+_20181908 | 3.02 |
ENSDART00000158987
|
hoxa4a
|
homeobox A4a |
| chr8_-_28415750 | 2.94 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr12_-_22438379 | 2.93 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr22_-_11712371 | 2.87 |
|
|
|
| chr19_-_47366517 | 2.84 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr19_+_20183210 | 2.65 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr3_+_23561215 | 2.65 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr23_+_28656263 | 2.59 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr12_-_10001043 | 2.52 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr14_-_5239605 | 2.51 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr4_+_5124186 | 2.46 |
ENSDART00000127874
|
ccnd2b
|
cyclin D2, b |
| chr11_+_43924640 | 2.43 |
ENSDART00000179206
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr22_-_16971799 | 2.42 |
ENSDART00000090242
|
nfia
|
nuclear factor I/A |
| chr14_+_30568242 | 2.25 |
|
|
|
| chr6_-_13000193 | 2.22 |
|
|
|
| chr15_-_25582891 | 2.20 |
|
|
|
| chr3_+_28871566 | 1.97 |
|
|
|
| chr25_+_34546629 | 1.92 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr22_-_16971035 | 1.88 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr22_-_29788268 | 1.88 |
|
|
|
| chr13_-_24316940 | 1.86 |
ENSDART00000127741
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr22_-_16971180 | 1.77 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr3_+_23561502 | 1.76 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr25_-_15117843 | 1.70 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr3_+_23561306 | 1.68 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr17_-_31204651 | 1.66 |
ENSDART00000137834
|
pcmtl
|
l-isoaspartyl protein carboxyl methyltransferase, like |
| chr20_+_53310252 | 1.65 |
ENSDART00000175125
|
CABZ01089116.1
|
ENSDARG00000106770 |
| chr20_+_32646617 | 1.63 |
|
|
|
| chr5_-_34393017 | 1.53 |
ENSDART00000134516
|
btf3
|
basic transcription factor 3 |
| chr25_-_20994084 | 1.44 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr6_-_40101104 | 1.41 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr1_+_18272833 | 1.41 |
ENSDART00000132379
|
limch1a
|
LIM and calponin homology domains 1a |
| chr11_-_3339651 | 1.38 |
|
|
|
| chr24_-_6129575 | 1.33 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr11_+_14373731 | 1.32 |
ENSDART00000161879
|
BX571942.1
|
ENSDARG00000099220 |
| chr22_+_24188575 | 1.32 |
ENSDART00000162227
|
glrx2
|
glutaredoxin 2 |
| chr15_+_16451044 | 1.24 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr11_+_6648811 | 1.22 |
ENSDART00000172061
|
CR381534.1
|
ENSDARG00000098564 |
| chr7_-_6331698 | 1.16 |
ENSDART00000166041
|
FP325123.2
|
Histone H3.2 |
| chr6_-_23193873 | 1.14 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr21_-_21111366 | 1.11 |
|
|
|
| chr19_+_20617339 | 1.08 |
ENSDART00000133633
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr10_+_42761097 | 0.96 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr9_-_33940350 | 0.92 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr9_+_1964320 | 0.92 |
|
|
|
| chr25_-_3766929 | 0.91 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr19_-_36009321 | 0.90 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr7_+_54336324 | 0.89 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
| chr10_+_37983342 | 0.89 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr7_-_26246473 | 0.86 |
ENSDART00000140694
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr16_-_36090863 | 0.85 |
|
|
|
| chr1_+_966385 | 0.82 |
ENSDART00000051919
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr2_-_39793399 | 0.82 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr8_-_24788837 | 0.80 |
ENSDART00000126220
|
alx3
|
ALX homeobox 3 |
| chr20_+_36098607 | 0.76 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr13_+_2213363 | 0.75 |
ENSDART00000019989
ENSDART00000165305 |
fbxo9
|
F-box protein 9 |
| chr5_-_18510716 | 0.71 |
ENSDART00000002624
ENSDART00000110553 |
ranbp1
|
RAN binding protein 1 |
| chr13_-_44601523 | 0.66 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr10_+_24721186 | 0.65 |
ENSDART00000144710
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr24_-_28150352 | 0.64 |
ENSDART00000145290
|
bcl2a
|
B-cell CLL/lymphoma 2a |
| chr25_+_35779830 | 0.60 |
ENSDART00000073405
|
zgc:173552
|
zgc:173552 |
| chr25_-_20993989 | 0.55 |
|
|
|
| chr16_+_22702982 | 0.55 |
ENSDART00000041625
|
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr9_-_33975936 | 0.55 |
ENSDART00000140779
|
fundc1
|
FUN14 domain containing 1 |
| chr21_+_17509116 | 0.51 |
|
|
|
| chr2_+_38165187 | 0.45 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr11_+_6138968 | 0.45 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr7_+_5783441 | 0.41 |
ENSDART00000167099
|
CU459186.5
|
Histone H3.2 |
| chr13_-_25643419 | 0.33 |
ENSDART00000145640
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr22_+_24188148 | 0.33 |
ENSDART00000167996
|
glrx2
|
glutaredoxin 2 |
| chr14_+_29933552 | 0.33 |
|
|
|
| chr13_+_24453943 | 0.33 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr9_+_41910191 | 0.33 |
|
|
|
| chr17_-_14697094 | 0.32 |
ENSDART00000154281
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr12_+_6161471 | 0.32 |
ENSDART00000163828
|
BX248126.1
|
ENSDARG00000096552 |
| chr18_+_14665460 | 0.24 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr17_+_13088982 | 0.22 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr15_+_27454643 | 0.20 |
ENSDART00000164887
ENSDART00000018603 |
tbx4
|
T-box 4 |
| chr3_+_33309011 | 0.17 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr25_+_3168376 | 0.16 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr2_-_55971357 | 0.14 |
ENSDART00000154701
ENSDART00000154107 |
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr7_+_5854890 | 0.10 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr25_-_35779675 | 0.10 |
|
|
|
| chr13_-_9543393 | 0.07 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr23_+_36020128 | 0.06 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr7_+_54336245 | 0.05 |
ENSDART00000168474
|
fgf3
|
fibroblast growth factor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.4 | 1.7 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 1.3 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.3 | 2.5 | GO:0048608 | gonad development(GO:0008406) negative regulation of endodermal cell fate specification(GO:0042664) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 2.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 1.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 6.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 0.9 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.2 | 3.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.2 | 0.8 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 3.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 5.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 2.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 | 0.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 2.5 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 1.0 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.7 | GO:0046607 | mitotic centrosome separation(GO:0007100) positive regulation of centrosome cycle(GO:0046607) centrosome separation(GO:0051299) |
| 0.1 | 0.4 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.6 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 1.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 15.4 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 5.7 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 5.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.9 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 4.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 6.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 5.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 6.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 3.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 4.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 11.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 3.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 2.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 1.6 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.3 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 2.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 1.0 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.2 | 0.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 2.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 5.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 4.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 31.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 6.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.9 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.2 | 2.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |