Project
DANIO-CODE
Navigation
Downloads
Results for hoxc4a
Z-value: 1.06
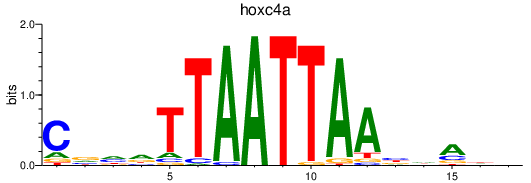
Transcription factors associated with hoxc4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc4a
|
ENSDARG00000070338 | homeobox C4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc4a | dr10_dc_chr23_+_36032206_36032259 | 0.93 | 1.8e-07 | Click! |
Activity profile of hoxc4a motif
Sorted Z-values of hoxc4a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc4a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23516127 | 4.63 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr3_-_32686790 | 4.48 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr16_-_42990753 | 3.84 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr8_+_21163236 | 3.54 |
ENSDART00000091307
|
col2a1a
|
collagen, type II, alpha 1a |
| chr20_-_9107294 | 3.37 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr19_+_10936229 | 3.24 |
ENSDART00000172219
|
apoea
|
apolipoprotein Ea |
| chr14_-_17257773 | 3.17 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr21_+_25199691 | 2.82 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr18_-_782643 | 2.79 |
ENSDART00000161084
|
si:dkey-205h23.1
|
si:dkey-205h23.1 |
| chr1_-_674449 | 2.66 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr15_-_14616083 | 2.55 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr16_+_46145286 | 2.53 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr19_+_43763483 | 2.26 |
ENSDART00000136695
|
yrk
|
Yes-related kinase |
| chr4_+_9668755 | 2.24 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr12_-_7902815 | 1.92 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr9_-_14533551 | 1.86 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr19_+_5562107 | 1.74 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr18_-_3056732 | 1.74 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr24_-_14447773 | 1.62 |
|
|
|
| chr25_-_5883620 | 1.57 |
ENSDART00000136054
|
snx22
|
sorting nexin 22 |
| chr23_+_7537089 | 1.47 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr2_-_49114158 | 1.47 |
|
|
|
| chr24_-_25546068 | 1.41 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr3_+_36155364 | 1.38 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr3_+_50511676 | 1.37 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr22_+_12746080 | 1.36 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr7_+_25052687 | 1.35 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr13_+_516287 | 1.35 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr2_+_51087570 | 1.34 |
|
|
|
| chr7_-_49322226 | 1.32 |
ENSDART00000174161
|
brsk2b
|
BR serine/threonine kinase 2b |
| chr23_-_7740845 | 1.32 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr2_+_3370130 | 1.29 |
ENSDART00000098391
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr24_+_21395671 | 1.23 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr14_+_2969679 | 1.22 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr17_-_26586506 | 1.20 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr21_+_26684617 | 1.18 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr13_-_49983998 | 1.17 |
|
|
|
| chr3_+_5665300 | 1.17 |
ENSDART00000101807
|
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr3_-_38642067 | 1.16 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr5_-_37850885 | 1.15 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr1_+_50395721 | 1.12 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr12_-_31679959 | 1.12 |
ENSDART00000153449
|
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr25_+_23326562 | 1.08 |
|
|
|
| chr10_-_11303185 | 1.04 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr18_-_43890514 | 0.98 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr24_-_25545773 | 0.96 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr5_+_11340748 | 0.93 |
ENSDART00000159248
|
tesca
|
tescalcin a |
| chr10_+_21693418 | 0.92 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr1_-_30801520 | 0.89 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr11_-_40414239 | 0.88 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr20_+_1724609 | 0.87 |
|
|
|
| chr25_+_23326630 | 0.82 |
|
|
|
| chr1_-_18809429 | 0.81 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr19_-_5453469 | 0.81 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr20_+_29307142 | 0.78 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr2_-_37061470 | 0.77 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr25_+_32114076 | 0.75 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr4_+_70768190 | 0.74 |
|
|
|
| chr1_-_9256864 | 0.74 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr8_+_29733109 | 0.73 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr5_+_45823481 | 0.71 |
|
|
|
| chr15_+_46813538 | 0.71 |
|
|
|
| chr3_-_15889742 | 0.70 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr8_-_12253026 | 0.68 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr17_+_42003279 | 0.68 |
ENSDART00000111537
|
kiz
|
kizuna centrosomal protein |
| chr22_+_699548 | 0.68 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr20_+_109649 | 0.67 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr24_-_28576494 | 0.66 |
|
|
|
| chr8_+_30654835 | 0.63 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr3_-_4111993 | 0.63 |
ENSDART00000170535
|
gucy2c
|
guanylate cyclase 2C |
| chr2_-_17721575 | 0.61 |
ENSDART00000141188
ENSDART00000100201 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr20_+_1724656 | 0.59 |
|
|
|
| chr24_+_35500964 | 0.59 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr24_-_14447136 | 0.59 |
|
|
|
| chr2_+_38390887 | 0.58 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr12_-_926641 | 0.58 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr20_+_29307039 | 0.58 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_+_36098607 | 0.56 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr22_-_12130441 | 0.54 |
ENSDART00000146785
|
tmem163b
|
transmembrane protein 163b |
| chr10_+_11303321 | 0.54 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr17_+_25425967 | 0.53 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr2_+_51087643 | 0.52 |
|
|
|
| chr2_-_37157969 | 0.52 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr24_+_24316486 | 0.52 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr17_+_44583892 | 0.52 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr13_+_4040744 | 0.51 |
|
|
|
| chr23_+_20714105 | 0.50 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr23_+_4773894 | 0.48 |
ENSDART00000172739
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr8_+_30654957 | 0.48 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr3_+_3994248 | 0.47 |
|
|
|
| chr3_-_13771322 | 0.46 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr3_+_27667194 | 0.44 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr16_+_54350976 | 0.42 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr7_+_21006803 | 0.41 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr2_+_50873893 | 0.38 |
|
|
|
| chr20_+_47639141 | 0.38 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr13_+_4040802 | 0.37 |
|
|
|
| chr12_+_17498203 | 0.36 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr8_+_30655100 | 0.35 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr22_+_1551406 | 0.35 |
ENSDART00000163223
|
si:ch211-255f4.9
|
si:ch211-255f4.9 |
| chr12_-_26062065 | 0.35 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr9_-_32366320 | 0.31 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr24_+_192889 | 0.31 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr2_+_37157979 | 0.28 |
ENSDART00000045016
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr6_+_40924749 | 0.28 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr7_-_950176 | 0.27 |
|
|
|
| chr13_+_516546 | 0.26 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr13_-_51743395 | 0.26 |
|
|
|
| chr17_-_20538582 | 0.24 |
|
|
|
| chr15_-_3024027 | 0.23 |
|
|
|
| chr24_+_24316408 | 0.23 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr21_+_6858020 | 0.23 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr22_+_12306260 | 0.22 |
|
|
|
| chr16_+_24818799 | 0.19 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr10_-_20545884 | 0.18 |
|
|
|
| chr21_+_26684728 | 0.18 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr12_+_1286911 | 0.17 |
ENSDART00000168225
ENSDART00000157467 |
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr20_-_43281081 | 0.16 |
|
|
|
| chr6_-_8156520 | 0.15 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr12_+_35755300 | 0.15 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr13_+_32609784 | 0.15 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr14_-_33604914 | 0.15 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr2_-_37061049 | 0.14 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_+_1881022 | 0.14 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr22_-_17571993 | 0.14 |
|
|
|
| chr13_+_516496 | 0.13 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr17_-_16414629 | 0.10 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr11_-_23333850 | 0.10 |
|
|
|
| chr24_+_24316346 | 0.10 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr16_+_40074850 | 0.10 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr7_-_52113622 | 0.08 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr7_+_24787720 | 0.06 |
|
|
|
| chr5_-_21015548 | 0.05 |
ENSDART00000040184
|
tenm1
|
teneurin transmembrane protein 1 |
| chr15_+_32940372 | 0.04 |
ENSDART00000170098
|
spg20b
|
spastic paraplegia 20b (Troyer syndrome) |
| chr12_+_48497255 | 0.04 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr19_+_46567459 | 0.04 |
ENSDART00000164055
|
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_57055662 | 0.04 |
ENSDART00000166545
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr18_-_15451738 | 0.03 |
ENSDART00000091466
|
polr3b
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr17_-_51804002 | 0.02 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0051000 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.5 | 1.4 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.4 | 1.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.3 | 3.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 1.5 | GO:0061469 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 1.9 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.2 | 1.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 | 1.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 3.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 2.3 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 1.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.8 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.3 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.5 | GO:0060753 | mast cell chemotaxis(GO:0002551) positive regulation of leukocyte chemotaxis(GO:0002690) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 2.4 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 2.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 3.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 3.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.4 | 3.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 4.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 1.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 1.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 3.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.6 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.6 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.0 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.0 | 0.5 | GO:0005254 | chloride channel activity(GO:0005254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 3.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |