Project
DANIO-CODE
Navigation
Downloads
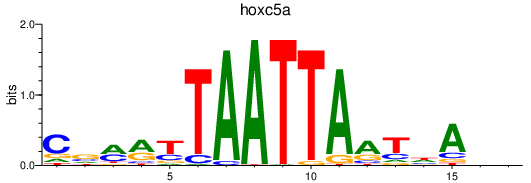
Results for hoxc5a
Z-value: 1.08
Transcription factors associated with hoxc5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc5a
|
ENSDARG00000070340 | homeobox C5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc5a | dr10_dc_chr23_+_36020128_36020168 | -0.74 | 1.1e-03 | Click! |
Activity profile of hoxc5a motif
Sorted Z-values of hoxc5a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc5a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_6289363 | 4.44 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| KN150456v1_-_19515 | 4.18 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr5_-_36503296 | 3.19 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr10_+_33449922 | 3.04 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr19_-_18664720 | 2.99 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr21_+_22598805 | 2.92 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr2_+_56534374 | 2.68 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| KN150699v1_-_15078 | 2.60 |
ENSDART00000159861
|
ENSDARG00000098739
|
ENSDARG00000098739 |
| chr12_+_47448318 | 2.49 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr8_-_32376710 | 2.28 |
ENSDART00000098850
|
lipg
|
lipase, endothelial |
| chr16_-_24727689 | 2.17 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr2_-_17721810 | 2.16 |
ENSDART00000100201
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr16_-_43061368 | 2.12 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr8_-_2557556 | 2.10 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr11_+_17849608 | 2.09 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr12_-_33256934 | 2.01 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr25_-_20933295 | 1.97 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr19_-_1571878 | 1.93 |
|
|
|
| chr18_+_39506453 | 1.89 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr12_+_33257120 | 1.88 |
|
|
|
| chr25_+_5845303 | 1.85 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr23_-_19299279 | 1.83 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr13_-_51743281 | 1.82 |
|
|
|
| chr12_-_33256754 | 1.79 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_+_8412489 | 1.78 |
ENSDART00000144851
|
zgc:113984
|
zgc:113984 |
| chr12_-_33257026 | 1.73 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr4_-_3340315 | 1.65 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr14_-_49777408 | 1.65 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr16_+_14075878 | 1.65 |
ENSDART00000059926
|
zgc:162509
|
zgc:162509 |
| chr24_+_19270877 | 1.62 |
|
|
|
| chr1_-_50395003 | 1.62 |
ENSDART00000035150
|
spast
|
spastin |
| chr19_-_18664670 | 1.60 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr11_+_23799984 | 1.56 |
|
|
|
| chr10_-_31129217 | 1.56 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr22_+_2735606 | 1.53 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr24_+_17115897 | 1.50 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr7_-_73016194 | 1.47 |
|
|
|
| chr24_-_9865837 | 1.46 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr18_-_43890836 | 1.45 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| KN149830v1_-_31870 | 1.45 |
ENSDART00000167653
|
zgc:194336
|
zgc:194336 |
| chr5_-_57053687 | 1.43 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr4_+_12967786 | 1.42 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr18_-_38787413 | 1.40 |
|
|
|
| chr25_-_20933145 | 1.40 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr14_-_15777250 | 1.40 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr13_-_17991273 | 1.39 |
ENSDART00000128748
|
fam21c
|
family with sequence similarity 21, member C |
| chr1_-_54570813 | 1.38 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr1_-_8244490 | 1.36 |
|
|
|
| chr21_+_40662814 | 1.35 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr5_-_3673698 | 1.34 |
|
|
|
| chr12_-_33256671 | 1.34 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_26675055 | 1.33 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr1_+_8468971 | 1.32 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr22_+_1335355 | 1.32 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr17_+_24300451 | 1.32 |
ENSDART00000064083
|
otx1b
|
orthodenticle homeobox 1b |
| chr21_+_27477153 | 1.31 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr3_-_39912816 | 1.30 |
ENSDART00000102540
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr25_+_32114026 | 1.29 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr24_+_26223767 | 1.28 |
|
|
|
| chr1_+_22160932 | 1.26 |
ENSDART00000016488
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr20_+_25687135 | 1.26 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr20_-_23977077 | 1.26 |
ENSDART00000043316
|
katna1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr7_+_66660882 | 1.26 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr6_-_50731449 | 1.24 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr10_-_17214368 | 1.24 |
|
|
|
| chr20_-_194135 | 1.24 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr21_+_13291305 | 1.24 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr23_-_19299398 | 1.23 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr4_+_4825461 | 1.22 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_-_29961498 | 1.22 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr18_-_16964811 | 1.21 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr20_-_29961589 | 1.21 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr18_+_35153749 | 1.20 |
ENSDART00000151073
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr10_-_31129270 | 1.19 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr2_-_31850133 | 1.19 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr15_+_34211736 | 1.18 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr25_-_13394261 | 1.16 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr7_-_26226924 | 1.15 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr3_-_40142513 | 1.14 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr19_-_25565317 | 1.14 |
ENSDART00000175266
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr10_+_1653720 | 1.14 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr6_-_10676857 | 1.13 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr15_+_35076414 | 1.13 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr16_+_42567707 | 1.13 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr1_+_58608231 | 1.13 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr21_+_10152870 | 1.13 |
|
|
|
| chr15_-_3023850 | 1.12 |
|
|
|
| chr7_-_73613531 | 1.12 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr6_+_18941186 | 1.11 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr19_+_1743359 | 1.11 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr1_+_157840 | 1.11 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr11_+_12754166 | 1.10 |
ENSDART00000123445
ENSDART00000163364 ENSDART00000066122 |
rtel1
|
regulator of telomere elongation helicase 1 |
| chr17_+_28085497 | 1.10 |
ENSDART00000131638
|
ENSDARG00000054122
|
ENSDARG00000054122 |
| chr2_-_21780380 | 1.09 |
ENSDART00000144587
|
plcd1b
|
phospholipase C, delta 1b |
| chr25_-_36616544 | 1.09 |
|
|
|
| chr20_-_47994260 | 1.09 |
ENSDART00000166857
|
CABZ01059120.1
|
ENSDARG00000100162 |
| chr20_-_35601497 | 1.09 |
|
|
|
| chr16_-_40633986 | 1.08 |
ENSDART00000170815
|
BX324007.2
|
ENSDARG00000100586 |
| chr6_+_342822 | 1.08 |
|
|
|
| chr19_-_25564720 | 1.07 |
ENSDART00000148432
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr20_+_2100836 | 1.06 |
|
|
|
| chr18_+_17736565 | 1.06 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr16_-_42015364 | 1.06 |
ENSDART00000136742
|
BX649388.1
|
ENSDARG00000091933 |
| chr19_-_5186692 | 1.05 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_+_57055662 | 1.05 |
ENSDART00000166545
|
fdxacb1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr5_-_31257158 | 1.05 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr21_+_13291242 | 1.05 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr8_-_53165501 | 1.05 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr3_+_52290252 | 1.04 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr13_+_40347099 | 1.04 |
|
|
|
| chr14_-_33141111 | 1.03 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr21_-_44086464 | 1.03 |
ENSDART00000130833
|
FO704810.1
|
ENSDARG00000087492 |
| chr5_-_31796734 | 1.02 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr25_-_31979408 | 1.02 |
|
|
|
| chr3_+_40022244 | 1.02 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr8_+_3364453 | 1.02 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr21_+_38697575 | 1.01 |
ENSDART00000113316
|
heatr6
|
HEAT repeat containing 6 |
| chr5_-_66145078 | 1.00 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr1_+_46808632 | 1.00 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr25_-_1243081 | 1.00 |
ENSDART00000156062
|
calml4b
|
calmodulin-like 4b |
| chr18_+_14715573 | 0.99 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr20_-_34126039 | 0.99 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr19_+_10742387 | 0.99 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr18_-_14306241 | 0.99 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr21_-_13026036 | 0.99 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr17_-_25813136 | 0.98 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr22_+_11943005 | 0.98 |
ENSDART00000105788
|
mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr11_+_30927094 | 0.98 |
ENSDART00000112098
|
stx10
|
syntaxin 10 |
| chr15_+_21327206 | 0.98 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr2_+_11094785 | 0.98 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr8_-_4517590 | 0.98 |
ENSDART00000090731
|
dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr8_+_29733037 | 0.98 |
ENSDART00000133955
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr5_-_36503236 | 0.97 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr3_+_12087549 | 0.97 |
|
|
|
| chr20_+_27341244 | 0.96 |
ENSDART00000123950
|
ENSDARG00000077377
|
ENSDARG00000077377 |
| chr11_+_25302044 | 0.96 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr7_+_35163845 | 0.95 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr17_-_43542320 | 0.95 |
ENSDART00000154595
|
si:dkey-21a6.5
|
si:dkey-21a6.5 |
| chr11_-_28508152 | 0.95 |
|
|
|
| chr11_+_11466531 | 0.95 |
ENSDART00000171536
|
Metazoa_SRP
|
Metazoan signal recognition particle RNA |
| chr9_+_2523927 | 0.94 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr6_+_18941289 | 0.93 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr2_-_19705537 | 0.93 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr21_-_39132388 | 0.93 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr13_-_50940520 | 0.92 |
|
|
|
| chr11_+_24562450 | 0.92 |
ENSDART00000082761
ENSDART00000131976 |
adipor1a
|
adiponectin receptor 1a |
| chr3_+_51308904 | 0.92 |
ENSDART00000008607
|
ttyh2l
|
tweety homolog 2, like |
| chr5_+_58883315 | 0.92 |
|
|
|
| chr23_-_25853364 | 0.92 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr18_+_15695287 | 0.92 |
|
|
|
| chr10_+_39390040 | 0.91 |
|
|
|
| chr11_+_11319810 | 0.91 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr7_-_73613763 | 0.91 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr15_+_34734212 | 0.90 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr23_-_41367783 | 0.90 |
|
|
|
| chr22_+_30041252 | 0.90 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr11_-_33956671 | 0.90 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr14_+_45274254 | 0.89 |
ENSDART00000176093
ENSDART00000175721 |
CR854988.1
|
ENSDARG00000105900 |
| chr2_-_11335644 | 0.89 |
ENSDART00000135450
|
cryz
|
crystallin, zeta (quinone reductase) |
| chr7_-_26226628 | 0.89 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr10_+_33450122 | 0.88 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr8_-_48679386 | 0.88 |
ENSDART00000047134
|
clcn6
|
chloride channel 6 |
| chr12_-_33256599 | 0.87 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_-_19890717 | 0.87 |
ENSDART00000104118
ENSDART00000170199 |
atxn7l3
|
ataxin 7-like 3 |
| chr20_-_3220106 | 0.86 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr15_-_18273640 | 0.86 |
ENSDART00000141508
|
btr16
|
bloodthirsty-related gene family, member 16 |
| chr15_+_25503446 | 0.85 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr15_+_45738963 | 0.85 |
|
|
|
| chr6_+_18941135 | 0.84 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr8_+_36527653 | 0.84 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr1_-_51075777 | 0.84 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr13_-_44694139 | 0.84 |
ENSDART00000137275
|
si:dkeyp-2e4.6
|
si:dkeyp-2e4.6 |
| chr21_+_13291347 | 0.84 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr4_-_62022037 | 0.83 |
|
|
|
| chr15_-_16241412 | 0.82 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr14_+_42109615 | 0.82 |
|
|
|
| chr10_+_1653868 | 0.82 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr3_+_35412519 | 0.82 |
ENSDART00000084549
|
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr22_-_16416848 | 0.81 |
ENSDART00000006290
|
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr24_-_28358595 | 0.81 |
ENSDART00000125412
|
fbxo45
|
F-box protein 45 |
| chr18_+_14651021 | 0.81 |
ENSDART00000166679
|
cirh1a
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr23_-_32173435 | 0.80 |
ENSDART00000044658
|
letmd1
|
LETM1 domain containing 1 |
| chr4_-_5100307 | 0.80 |
ENSDART00000138817
|
tmem209
|
transmembrane protein 209 |
| chr14_-_38942 | 0.80 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr14_-_44880144 | 0.79 |
ENSDART00000163543
|
abhd18
|
abhydrolase domain containing 18 |
| chr16_-_42015262 | 0.79 |
ENSDART00000136742
|
BX649388.1
|
ENSDARG00000091933 |
| chr1_+_58608185 | 0.79 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr11_+_25302180 | 0.79 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr14_+_47229149 | 0.79 |
ENSDART00000136045
|
mgst2
|
microsomal glutathione S-transferase 2 |
| chr5_-_40503137 | 0.78 |
ENSDART00000051070
|
golph3
|
golgi phosphoprotein 3 |
| chr13_+_9487120 | 0.78 |
ENSDART00000091595
|
fam45a
|
family with sequence similarity 45, member A |
| chr6_+_49552893 | 0.78 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr13_+_12538980 | 0.78 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr22_-_28276824 | 0.78 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr8_+_30103357 | 0.77 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr21_+_40663230 | 0.77 |
ENSDART00000017709
|
ccdc82
|
coiled-coil domain containing 82 |
| chr5_+_53851006 | 0.76 |
ENSDART00000157722
ENSDART00000160446 |
tmem203
|
transmembrane protein 203 |
| chr22_-_23252993 | 0.76 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr7_-_54752916 | 0.75 |
|
|
|
| chr5_+_25133592 | 0.75 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.7 | 2.8 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.5 | 3.9 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.5 | 1.9 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.4 | 1.6 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.4 | 2.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.4 | 1.1 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.4 | 1.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) temperature homeostasis(GO:0001659) |
| 0.3 | 1.0 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 0.9 | GO:0070655 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.3 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 0.8 | GO:2000106 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.3 | 1.6 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.3 | 1.0 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 0.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 2.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 0.6 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.6 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 0.6 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 0.7 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.2 | 0.5 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 2.4 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 0.8 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 7.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.2 | 1.5 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.2 | 0.5 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.2 | 1.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 4.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 3.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.7 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.1 | 0.7 | GO:0046320 | regulation of fatty acid beta-oxidation(GO:0031998) regulation of fatty acid oxidation(GO:0046320) |
| 0.1 | 2.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.4 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.8 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 0.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.3 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.6 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.6 | GO:0045761 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 0.4 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 1.6 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 0.9 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.9 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 1.0 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.3 | GO:0071498 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 1.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.6 | GO:0097345 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.7 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.3 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.5 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 1.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 1.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 2.0 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 1.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.7 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 1.3 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 2.5 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.1 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 1.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 1.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.6 | GO:0001878 | response to yeast(GO:0001878) response to fungus(GO:0009620) |
| 0.0 | 0.1 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.0 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 2.4 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.3 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.1 | GO:0009060 | aerobic respiration(GO:0009060) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.4 | 2.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 1.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 2.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.3 | 1.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 1.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 1.0 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.2 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 2.4 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 6.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.4 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 1.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.3 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 5.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 5.7 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 3.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.3 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.7 | 2.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.6 | 1.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.5 | 1.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.4 | 2.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 1.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.4 | 2.4 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.4 | 1.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.4 | 2.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 1.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.1 | GO:0070042 | rRNA (uridine-N3-)-methyltransferase activity(GO:0070042) |
| 0.3 | 1.2 | GO:0019705 | protein-cysteine S-myristoyltransferase activity(GO:0019705) |
| 0.3 | 3.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 0.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 0.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 2.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.3 | 1.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.7 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 3.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.6 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 2.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 3.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 1.0 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 1.0 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 4.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.9 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 3.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.2 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.5 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.9 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 2.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 1.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0019869 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 8.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 2.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 1.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 2.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.6 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |