Project
DANIO-CODE
Navigation
Downloads
Results for hoxc6a+hoxc6b
Z-value: 0.79
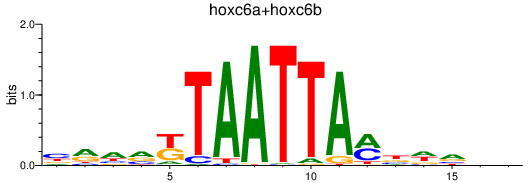
Transcription factors associated with hoxc6a+hoxc6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc6a
|
ENSDARG00000070343 | homeobox C6a |
|
hoxc6b
|
ENSDARG00000101954 | homeobox C6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc6b | dr10_dc_chr11_-_2107054_2107087 | 0.42 | 1.0e-01 | Click! |
| hoxc6a | dr10_dc_chr23_+_36016450_36016460 | 0.22 | 4.2e-01 | Click! |
Activity profile of hoxc6a+hoxc6b motif
Sorted Z-values of hoxc6a+hoxc6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc6a+hoxc6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_4825019 | 2.14 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr3_-_19218660 | 1.61 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr18_+_40365208 | 1.48 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr1_-_674449 | 1.40 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr3_+_30563531 | 1.27 |
ENSDART00000153958
ENSDART00000154331 |
BX682556.1
|
ENSDARG00000097595 |
| chr19_-_5851328 | 1.23 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr10_-_15896595 | 1.18 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr6_+_35378839 | 1.09 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr12_-_4497094 | 1.07 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr4_-_73485204 | 1.04 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr24_+_34227202 | 1.00 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr20_-_45908435 | 0.91 |
ENSDART00000147637
|
fermt1
|
fermitin family member 1 |
| chr9_-_8035360 | 0.90 |
|
|
|
| chr21_+_25199691 | 0.88 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr19_-_27379989 | 0.88 |
ENSDART00000103955
|
zbtb12.2
|
zinc finger and BTB domain containing 12, tandem duplicate 2 |
| chr10_-_43924675 | 0.84 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr21_-_32267773 | 0.84 |
ENSDART00000134414
|
clk4b
|
CDC-like kinase 4b |
| chr3_+_22962233 | 0.84 |
ENSDART00000142884
|
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr20_+_109649 | 0.83 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr24_+_35500964 | 0.83 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr6_-_343209 | 0.80 |
|
|
|
| chr10_-_7427767 | 0.80 |
ENSDART00000167963
ENSDART00000162021 |
nrg1
|
neuregulin 1 |
| chr22_-_36561247 | 0.74 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr15_+_42329269 | 0.71 |
ENSDART00000099234
ENSDART00000152731 |
scaf4b
|
SR-related CTD-associated factor 4b |
| chr10_+_11809550 | 0.68 |
ENSDART00000092047
|
ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr24_+_21395671 | 0.67 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr20_-_9107294 | 0.66 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr6_-_53227526 | 0.65 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr23_+_29431734 | 0.64 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr13_+_35213326 | 0.62 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr5_-_9162684 | 0.62 |
|
|
|
| chr10_+_7125001 | 0.61 |
ENSDART00000157987
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr1_+_50395721 | 0.61 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr14_-_41751299 | 0.60 |
ENSDART00000128360
|
BX537109.1
|
ENSDARG00000086015 |
| chr4_-_16555765 | 0.59 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr17_-_32471070 | 0.58 |
|
|
|
| chr17_+_30352361 | 0.56 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr3_-_31027448 | 0.54 |
ENSDART00000076796
|
smg1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr12_-_28248133 | 0.53 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr13_+_516287 | 0.53 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr7_+_58718614 | 0.51 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr16_-_21334269 | 0.50 |
ENSDART00000145837
|
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr10_-_11303185 | 0.48 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr12_+_27040886 | 0.47 |
|
|
|
| chr13_-_51743395 | 0.45 |
|
|
|
| chr7_+_7463292 | 0.44 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr2_+_1825714 | 0.43 |
ENSDART00000148624
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr7_-_26331734 | 0.42 |
|
|
|
| chr21_-_26453406 | 0.41 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr2_+_20748431 | 0.40 |
ENSDART00000137848
|
palmda
|
palmdelphin a |
| chr9_-_32366320 | 0.39 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr10_+_11303321 | 0.37 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr20_-_35343057 | 0.37 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr16_-_27743294 | 0.36 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr19_+_43763483 | 0.36 |
ENSDART00000136695
|
yrk
|
Yes-related kinase |
| chr22_-_27002690 | 0.35 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr16_+_13928844 | 0.35 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr21_-_36886505 | 0.35 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr2_+_3370130 | 0.34 |
ENSDART00000098391
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr13_+_1377875 | 0.33 |
|
|
|
| chr7_-_5253397 | 0.33 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr4_-_73485366 | 0.33 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr3_-_13771322 | 0.33 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr18_+_7607055 | 0.33 |
ENSDART00000140784
|
CR318588.4
|
ENSDARG00000095556 |
| chr16_-_7876036 | 0.32 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr5_+_45823481 | 0.32 |
|
|
|
| chr4_-_28675011 | 0.31 |
ENSDART00000163985
|
CABZ01050665.1
|
ENSDARG00000104281 |
| chr14_-_6831308 | 0.30 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr13_+_516546 | 0.30 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr10_-_20545884 | 0.30 |
|
|
|
| chr17_+_42003279 | 0.30 |
ENSDART00000111537
|
kiz
|
kizuna centrosomal protein |
| chr22_+_1551406 | 0.29 |
ENSDART00000163223
|
si:ch211-255f4.9
|
si:ch211-255f4.9 |
| chr4_+_9668755 | 0.29 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr22_+_470536 | 0.29 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr5_-_12751614 | 0.28 |
ENSDART00000030553
|
top3b
|
topoisomerase (DNA) III beta |
| chr16_+_54350976 | 0.28 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr4_-_44665935 | 0.28 |
ENSDART00000150524
|
si:ch211-162i8.4
|
si:ch211-162i8.4 |
| chr20_+_25326042 | 0.27 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
| chr6_-_16590883 | 0.27 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr17_+_25500291 | 0.27 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr25_+_23326562 | 0.27 |
|
|
|
| chr20_-_54206786 | 0.27 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr6_+_7376428 | 0.26 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr10_+_29964045 | 0.26 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr15_-_26998580 | 0.26 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr17_+_43605118 | 0.24 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr13_+_516496 | 0.24 |
ENSDART00000172469
ENSDART00000168523 ENSDART00000006892 |
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr21_+_45802046 | 0.24 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr14_-_6831373 | 0.24 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr7_-_50137683 | 0.23 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr3_+_28808901 | 0.22 |
ENSDART00000141904
ENSDART00000077221 |
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr17_-_16414629 | 0.21 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr25_+_35771748 | 0.21 |
|
|
|
| chr22_+_19615230 | 0.20 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr11_-_40464213 | 0.20 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr22_-_16128314 | 0.20 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr2_-_17564676 | 0.19 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr7_+_19243998 | 0.19 |
ENSDART00000173766
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr6_+_14854101 | 0.19 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr9_-_32803650 | 0.18 |
|
|
|
| chr22_+_12306260 | 0.18 |
|
|
|
| chr18_-_43890514 | 0.17 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr8_-_19166630 | 0.17 |
|
|
|
| chr4_-_73485324 | 0.17 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr15_-_2790691 | 0.16 |
ENSDART00000102096
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr21_-_27584637 | 0.16 |
|
|
|
| chr12_-_41128584 | 0.16 |
|
|
|
| chr10_+_29964164 | 0.16 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr22_+_20141311 | 0.15 |
|
|
|
| chr1_-_22160662 | 0.15 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr10_-_35714800 | 0.15 |
|
|
|
| chr11_+_36983626 | 0.15 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr15_+_42329433 | 0.15 |
ENSDART00000130404
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr8_+_36527535 | 0.15 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr23_+_36016450 | 0.14 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr10_+_29963998 | 0.13 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr7_-_5366526 | 0.13 |
|
|
|
| chr14_-_17000025 | 0.13 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr8_-_32497168 | 0.13 |
ENSDART00000164826
|
RFESD (1 of many)
|
Rieske Fe-S domain containing |
| chr11_-_6442836 | 0.12 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr20_+_22900165 | 0.12 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr4_+_2551928 | 0.12 |
ENSDART00000103371
ENSDART00000171405 |
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr12_+_1286911 | 0.11 |
ENSDART00000168225
ENSDART00000157467 |
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr5_-_68712571 | 0.11 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr20_+_22900302 | 0.10 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr25_+_23326630 | 0.10 |
|
|
|
| chr8_+_26336302 | 0.09 |
|
|
|
| chr7_+_7463565 | 0.09 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr6_+_24299180 | 0.09 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr2_-_53156364 | 0.08 |
|
|
|
| chr16_+_40074850 | 0.08 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr8_+_17479651 | 0.07 |
|
|
|
| chr23_-_35691396 | 0.07 |
ENSDART00000011004
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr25_-_20933145 | 0.07 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr15_-_43402935 | 0.07 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr5_-_68712851 | 0.07 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr2_-_55565099 | 0.07 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr19_+_5562107 | 0.07 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr25_-_27279272 | 0.06 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr5_-_42304154 | 0.06 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr14_-_38441710 | 0.06 |
|
|
|
| chr25_+_1046304 | 0.05 |
ENSDART00000172662
|
fam96a
|
family with sequence similarity 96, member A |
| chr22_-_4867538 | 0.04 |
ENSDART00000081801
|
ncl1
|
nicalin |
| chr11_-_40464253 | 0.04 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr1_+_56554227 | 0.04 |
ENSDART00000149688
|
mycbpap
|
mycbp associated protein |
| chr25_+_10450395 | 0.03 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr9_+_55395276 | 0.03 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr15_+_9351211 | 0.03 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr15_-_36490729 | 0.02 |
|
|
|
| chr11_+_17849528 | 0.02 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr20_-_15029379 | 0.01 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr14_+_37317006 | 0.01 |
|
|
|
| chr20_+_36098607 | 0.01 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr3_-_51038166 | 0.01 |
|
|
|
| chr2_-_37157969 | 0.00 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.3 | 0.8 | GO:0021755 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.7 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 1.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.3 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.3 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.1 | 0.3 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.3 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.1 | 0.3 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.7 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:1904407 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 1.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 1.1 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.2 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 0.5 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 0.7 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.2 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.5 | GO:0000215 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |