Project
DANIO-CODE
Navigation
Downloads
Results for hoxc8a
Z-value: 0.65
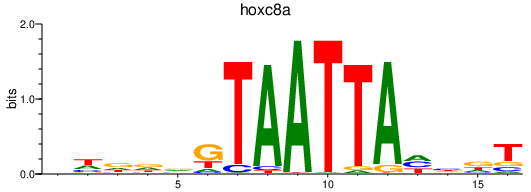
Transcription factors associated with hoxc8a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc8a
|
ENSDARG00000070346 | homeobox C8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc8a | dr10_dc_chr23_+_36002332_36002431 | 0.50 | 4.8e-02 | Click! |
Activity profile of hoxc8a motif
Sorted Z-values of hoxc8a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc8a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_57300743 | 1.82 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr2_+_4366169 | 1.41 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr13_+_35213326 | 1.35 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr4_-_5325164 | 1.30 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr20_-_45908435 | 1.28 |
ENSDART00000147637
|
fermt1
|
fermitin family member 1 |
| chr13_-_33574216 | 1.25 |
ENSDART00000065435
|
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr17_+_23278879 | 1.16 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr5_+_27897765 | 1.06 |
ENSDART00000166737
ENSDART00000164418 |
nfr
|
notochord formation related |
| chr12_-_4497094 | 1.04 |
ENSDART00000163651
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr18_-_3056732 | 1.02 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr21_-_22510751 | 1.00 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr4_-_22751641 | 0.97 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr10_+_42530040 | 0.89 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr15_+_880573 | 0.86 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr16_+_51316772 | 0.85 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr1_-_40208544 | 0.80 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr20_-_9107294 | 0.77 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr15_-_15421065 | 0.74 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr23_+_36032206 | 0.69 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr15_-_14616083 | 0.68 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr10_+_38582701 | 0.67 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr9_-_47833777 | 0.62 |
|
|
|
| chr16_-_13733759 | 0.61 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr1_-_40208469 | 0.59 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr2_+_4366568 | 0.58 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr8_-_13026629 | 0.57 |
ENSDART00000129168
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr17_+_30352361 | 0.57 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr8_-_31098193 | 0.57 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr2_-_17564676 | 0.55 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr15_-_35394535 | 0.55 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr19_-_39048324 | 0.54 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr15_-_1858350 | 0.53 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr4_-_76594399 | 0.51 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr4_+_14728248 | 0.50 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr8_+_18593622 | 0.50 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr16_-_13734068 | 0.50 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr25_+_4918339 | 0.48 |
ENSDART00000153980
|
parvb
|
parvin, beta |
| chr21_-_26453406 | 0.48 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr11_-_40464213 | 0.47 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr1_-_57301027 | 0.46 |
ENSDART00000081122
|
COLGALT1 (1 of many)
|
collagen beta(1-O)galactosyltransferase 1 |
| chr7_-_26331734 | 0.45 |
|
|
|
| chr6_+_24299180 | 0.45 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr23_-_7740845 | 0.44 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr15_-_34987450 | 0.44 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr6_-_53227526 | 0.44 |
ENSDART00000160955
|
slc38a3b
|
solute carrier family 38, member 3b |
| chr8_-_19166630 | 0.42 |
|
|
|
| chr5_-_27958815 | 0.42 |
ENSDART00000177313
|
si:ch73-195i19.3
|
si:ch73-195i19.3 |
| chr19_-_39048402 | 0.41 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr20_+_38298755 | 0.40 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr2_+_4366348 | 0.39 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr1_-_518543 | 0.39 |
ENSDART00000138032
|
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr2_-_28687065 | 0.38 |
ENSDART00000164657
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr24_-_25283218 | 0.37 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr5_+_65754237 | 0.36 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr4_-_802415 | 0.35 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr7_-_45726932 | 0.35 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr12_+_18622682 | 0.34 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr5_+_62987426 | 0.34 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr20_+_38298893 | 0.34 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr2_-_53156364 | 0.33 |
|
|
|
| chr5_+_29071526 | 0.33 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr1_+_22000499 | 0.33 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr7_-_23775835 | 0.32 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr10_-_5016472 | 0.31 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr20_+_41123859 | 0.31 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr2_+_4366515 | 0.31 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr7_-_45726974 | 0.31 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr6_-_8075384 | 0.30 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr8_-_12365342 | 0.29 |
ENSDART00000113286
|
phf19
|
PHD finger protein 19 |
| chr3_+_27667194 | 0.28 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr13_+_13562712 | 0.28 |
ENSDART00000110509
ENSDART00000169265 |
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr10_+_22065010 | 0.28 |
ENSDART00000133304
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr15_-_5911548 | 0.28 |
|
|
|
| chr23_+_36016450 | 0.27 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr8_-_7548976 | 0.27 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr1_+_18610282 | 0.27 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr21_+_35292807 | 0.27 |
ENSDART00000130626
|
rars
|
arginyl-tRNA synthetase |
| chr6_+_33896684 | 0.26 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr18_-_510172 | 0.25 |
ENSDART00000164374
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr21_-_11561978 | 0.25 |
ENSDART00000132699
|
cast
|
calpastatin |
| chr25_+_29690047 | 0.25 |
|
|
|
| chr11_-_40464253 | 0.24 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr17_+_25500291 | 0.24 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr21_-_5692160 | 0.24 |
ENSDART00000075137
ENSDART00000151202 |
ccni
|
cyclin I |
| chr19_+_5562107 | 0.23 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr10_-_20545884 | 0.23 |
|
|
|
| chr8_-_12365285 | 0.23 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr19_-_34263023 | 0.22 |
ENSDART00000160719
|
otud6b
|
OTU domain containing 6B |
| chr14_-_3836967 | 0.20 |
ENSDART00000055817
|
ENSDARG00000038270
|
ENSDARG00000038270 |
| chr9_+_41278246 | 0.19 |
ENSDART00000141179
|
zgc:136439
|
zgc:136439 |
| chr20_+_13507202 | 0.19 |
ENSDART00000127350
|
enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr2_+_50873893 | 0.18 |
|
|
|
| chr20_-_15029379 | 0.18 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr15_-_5911817 | 0.16 |
|
|
|
| chr20_-_16553010 | 0.16 |
|
|
|
| chr15_-_18130992 | 0.16 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr15_-_5912105 | 0.15 |
|
|
|
| chr9_+_4708975 | 0.14 |
ENSDART00000081326
|
prpf40a
|
PRP40 pre-mRNA processing factor 40 homolog A |
| chr2_-_32842678 | 0.13 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr15_-_37202518 | 0.13 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr20_-_9474672 | 0.12 |
ENSDART00000152674
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr2_+_4366022 | 0.12 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr11_+_11284030 | 0.11 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr22_-_16128314 | 0.11 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr9_+_32367808 | 0.11 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr3_+_19548039 | 0.10 |
|
|
|
| chr23_-_35691396 | 0.10 |
ENSDART00000011004
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr4_+_5325362 | 0.09 |
ENSDART00000150306
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr8_+_28362311 | 0.09 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr10_+_11397605 | 0.08 |
ENSDART00000064215
|
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr10_+_6925975 | 0.08 |
|
|
|
| chr12_-_36303384 | 0.08 |
ENSDART00000168538
|
oxld1
|
oxidoreductase-like domain containing 1 |
| chr15_-_36072087 | 0.07 |
ENSDART00000076229
|
irs1
|
insulin receptor substrate 1 |
| chr25_-_3345176 | 0.06 |
ENSDART00000029067
|
hbp1
|
HMG-box transcription factor 1 |
| chr5_-_42304154 | 0.06 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr15_-_5911297 | 0.06 |
|
|
|
| chr17_-_14621328 | 0.05 |
|
|
|
| chr15_+_22331719 | 0.05 |
ENSDART00000110665
|
nrgnb
|
neurogranin (protein kinase C substrate, RC3) b |
| chr13_+_6091028 | 0.05 |
ENSDART00000025910
|
apmap
|
adipocyte plasma membrane associated protein |
| chr3_-_40407003 | 0.03 |
|
|
|
| chr21_+_29040831 | 0.02 |
ENSDART00000166403
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr20_-_49901709 | 0.01 |
ENSDART00000129214
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr2_-_33703921 | 0.01 |
ENSDART00000147439
|
atp6v0b
|
ATPase, H+ transporting, lysosomal V0 subunit b |
| chr23_+_29431734 | 0.01 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr15_-_37202326 | 0.01 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr3_+_22312446 | 0.00 |
ENSDART00000156450
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 | 2.8 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.3 | 1.0 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.3 | 1.0 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.9 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.7 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.3 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.0 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.2 | GO:0055062 | regulation of fat cell differentiation(GO:0045598) phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.4 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.4 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.6 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0030677 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 0.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.3 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 1.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |