Project
DANIO-CODE
Navigation
Downloads
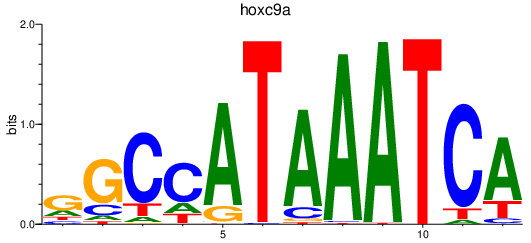
Results for hoxc9a
Z-value: 0.84
Transcription factors associated with hoxc9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc9a
|
ENSDARG00000092809 | homeobox C9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc9a | dr10_dc_chr23_+_35996491_35996540 | 0.86 | 2.2e-05 | Click! |
Activity profile of hoxc9a motif
Sorted Z-values of hoxc9a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc9a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_55022668 | 3.00 |
ENSDART00000158845
|
mybphb
|
myosin binding protein Hb |
| chr3_-_60973097 | 2.94 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr5_-_50972677 | 2.76 |
ENSDART00000134606
|
otpb
|
orthopedia homeobox b |
| chr9_-_1954910 | 2.70 |
ENSDART00000140438
|
BX322661.1
|
ENSDARG00000094186 |
| chr8_+_47644421 | 2.66 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr2_-_36058327 | 1.95 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr8_+_15968469 | 1.94 |
ENSDART00000134787
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_+_20183210 | 1.92 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr11_+_15905877 | 1.82 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr10_+_10830549 | 1.80 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr20_-_39003917 | 1.76 |
|
|
|
| chr5_-_50972592 | 1.76 |
ENSDART00000134606
|
otpb
|
orthopedia homeobox b |
| chr3_+_33168814 | 1.73 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr10_-_10374032 | 1.68 |
ENSDART00000134929
|
BX323065.1
|
ENSDARG00000095365 |
| chr22_-_21021645 | 1.60 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr3_-_6078015 | 1.55 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr21_-_1716495 | 1.44 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr19_-_39048324 | 1.36 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr19_+_20202737 | 1.34 |
ENSDART00000164677
|
hoxa4a
|
homeobox A4a |
| chr17_+_32390603 | 1.34 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr20_-_39003972 | 1.32 |
|
|
|
| chr19_+_20181908 | 1.31 |
ENSDART00000158987
|
hoxa4a
|
homeobox A4a |
| chr8_+_6533379 | 1.26 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr19_+_43414160 | 1.26 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr5_+_15880920 | 1.23 |
ENSDART00000168643
|
CABZ01088700.1
|
ENSDARG00000101452 |
| chr3_-_34363698 | 1.20 |
|
|
|
| chr13_+_14873339 | 1.19 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| KN150001v1_+_14939 | 1.15 |
|
|
|
| chr7_+_31608828 | 1.08 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_40667131 | 1.01 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr25_+_2933235 | 0.98 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| KN150207v1_-_1208 | 0.97 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr9_-_40212864 | 0.95 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr13_-_43466076 | 0.95 |
ENSDART00000172636
ENSDART00000084416 |
ablim1a
|
actin binding LIM protein 1a |
| chr8_+_39525254 | 0.93 |
|
|
|
| chr21_+_13063614 | 0.92 |
|
|
|
| chr4_+_10016118 | 0.91 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr2_-_41873625 | 0.87 |
ENSDART00000155577
|
ENSDARG00000056490
|
ENSDARG00000056490 |
| chr23_-_24240728 | 0.86 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr7_-_68417190 | 0.81 |
|
|
|
| chr10_+_13042496 | 0.81 |
ENSDART00000158919
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr5_-_22847399 | 0.80 |
ENSDART00000169258
|
nlgn3b
|
neuroligin 3b |
| chr9_-_40212643 | 0.80 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr9_-_3700395 | 0.78 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr15_-_16075172 | 0.78 |
|
|
|
| chr3_-_37558102 | 0.73 |
ENSDART00000151810
|
gpatch8
|
G patch domain containing 8 |
| chr23_-_4175790 | 0.73 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr19_+_27895343 | 0.70 |
ENSDART00000049368
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr23_+_35980812 | 0.68 |
|
|
|
| chr10_+_26609369 | 0.68 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr21_-_27301938 | 0.67 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr11_+_18710724 | 0.63 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr15_-_510285 | 0.62 |
ENSDART00000058993
|
psph
|
phosphoserine phosphatase |
| chr1_-_22466834 | 0.60 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr19_-_39048402 | 0.58 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr17_-_19606453 | 0.57 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr12_-_16331781 | 0.55 |
ENSDART00000152133
|
hectd2
|
HECT domain containing 2 |
| chr9_-_40213039 | 0.51 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr22_-_21021866 | 0.50 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr12_+_27122352 | 0.50 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr3_+_23608395 | 0.46 |
|
|
|
| chr15_-_14439483 | 0.45 |
ENSDART00000160675
|
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr23_-_24240798 | 0.45 |
ENSDART00000135173
|
CR925709.1
|
ENSDARG00000092130 |
| chr1_+_33510660 | 0.44 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr5_-_64637049 | 0.43 |
ENSDART00000164649
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr7_-_26137243 | 0.43 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr19_-_31378033 | 0.42 |
|
|
|
| chr18_+_13589341 | 0.41 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr12_+_16391933 | 0.34 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr22_+_35113233 | 0.33 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| KN150001v1_+_15017 | 0.33 |
|
|
|
| chr15_+_8791540 | 0.32 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr24_-_32115473 | 0.32 |
ENSDART00000168419
|
rsu1
|
Ras suppressor protein 1 |
| chr9_-_40213104 | 0.31 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr2_+_1881022 | 0.31 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr15_+_29241472 | 0.29 |
|
|
|
| chr9_+_492585 | 0.28 |
ENSDART00000112635
|
ENSDARG00000078172
|
ENSDARG00000078172 |
| chr25_+_32419062 | 0.28 |
|
|
|
| chr16_+_42161483 | 0.26 |
|
|
|
| chr17_+_51173489 | 0.23 |
ENSDART00000063738
|
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr5_-_62187774 | 0.22 |
|
|
|
| chr17_-_51173358 | 0.20 |
ENSDART00000169050
|
trappc12
|
trafficking protein particle complex 12 |
| chr9_+_44921233 | 0.20 |
ENSDART00000145271
|
nckap1
|
NCK-associated protein 1 |
| chr7_+_30779987 | 0.20 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr14_+_11457009 | 0.19 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr23_+_8862155 | 0.18 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr3_+_40667088 | 0.17 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr1_-_686007 | 0.11 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr9_+_1964320 | 0.11 |
|
|
|
| chr21_-_11877044 | 0.08 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr14_-_25645760 | 0.07 |
ENSDART00000141916
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr11_-_16261123 | 0.06 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr4_-_18646081 | 0.04 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr16_+_50489655 | 0.03 |
|
|
|
| chr24_-_21740552 | 0.02 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.9 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.3 | 1.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 0.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 1.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.2 | 1.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.2 | 0.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.6 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.7 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 1.8 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.3 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.9 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.3 | GO:0046958 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 1.0 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.7 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 4.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 2.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.8 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 1.3 | GO:0001947 | heart looping(GO:0001947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) ferric iron binding(GO:0008199) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.9 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |