Project
DANIO-CODE
Navigation
Downloads
Results for hoxd10a
Z-value: 0.39
Transcription factors associated with hoxd10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd10a
|
ENSDARG00000057859 | homeobox D10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd10a | dr10_dc_chr9_-_1969197_1969282 | -0.61 | 1.2e-02 | Click! |
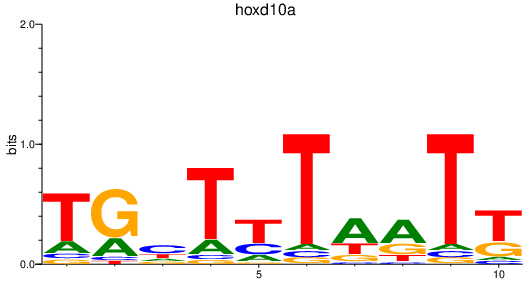
Activity profile of hoxd10a motif
Sorted Z-values of hoxd10a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd10a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_2373601 | 0.77 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr19_+_14132374 | 0.61 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr19_+_14132473 | 0.55 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr8_-_28359022 | 0.48 |
ENSDART00000062693
|
ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr17_+_31605159 | 0.48 |
ENSDART00000157148
|
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr9_-_3548174 | 0.47 |
ENSDART00000145043
ENSDART00000169586 |
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr25_+_18339864 | 0.47 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr16_-_42236126 | 0.46 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr21_+_21242470 | 0.45 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr12_+_30673985 | 0.45 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_+_9579430 | 0.43 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr6_-_39634534 | 0.43 |
ENSDART00000077839
|
atf7b
|
activating transcription factor 7b |
| chr6_-_25065823 | 0.42 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr6_-_25065745 | 0.37 |
ENSDART00000165170
|
znf326
|
zinc finger protein 326 |
| chr15_-_5913264 | 0.35 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr16_+_17857068 | 0.35 |
ENSDART00000149408
|
them4
|
thioesterase superfamily member 4 |
| chr22_-_11024649 | 0.34 |
ENSDART00000105823
ENSDART00000159995 |
insrb
|
insulin receptor b |
| chr9_-_3548211 | 0.33 |
ENSDART00000145043
ENSDART00000169586 |
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr6_-_39524075 | 0.33 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr25_-_3091345 | 0.32 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr5_+_30386444 | 0.32 |
ENSDART00000005893
|
ube2g1a
|
ubiquitin-conjugating enzyme E2G 1a (UBC7 homolog, yeast) |
| chr14_+_44343015 | 0.30 |
ENSDART00000079866
|
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr7_-_18624798 | 0.30 |
ENSDART00000113593
|
mllt3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr5_-_35997345 | 0.30 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr11_+_30005716 | 0.30 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr13_+_28481835 | 0.30 |
ENSDART00000077383
|
borcs7
|
BLOC-1 related complex subunit 7 |
| chr6_-_39634666 | 0.28 |
ENSDART00000077839
|
atf7b
|
activating transcription factor 7b |
| chr23_-_41367783 | 0.28 |
|
|
|
| chr11_-_38856849 | 0.28 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr22_-_22277418 | 0.28 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr3_-_15346427 | 0.28 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr3_-_26060787 | 0.26 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr17_+_20231630 | 0.26 |
ENSDART00000155584
|
BX005375.1
|
ENSDARG00000097982 |
| chr3_-_54414615 | 0.26 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr18_-_199009 | 0.26 |
|
|
|
| chr25_+_18339837 | 0.23 |
ENSDART00000178154
|
cep41
|
centrosomal protein 41 |
| chr2_+_21698414 | 0.23 |
ENSDART00000171397
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr2_-_49297561 | 0.21 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr11_-_10472565 | 0.20 |
ENSDART00000011087
|
ect2
|
epithelial cell transforming 2 |
| chr9_+_3547661 | 0.20 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr20_-_13244237 | 0.19 |
ENSDART00000124470
|
ints7
|
integrator complex subunit 7 |
| chr11_-_18112090 | 0.19 |
ENSDART00000019248
|
tmem110
|
transmembrane protein 110 |
| chr18_-_20469413 | 0.19 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr13_+_49436597 | 0.19 |
ENSDART00000037559
ENSDART00000168799 ENSDART00000097845 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr1_-_17018695 | 0.18 |
ENSDART00000148322
|
ufsp2
|
ufm1-specific peptidase 2 |
| chr21_+_3645203 | 0.18 |
ENSDART00000099535
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr3_-_26060702 | 0.18 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr7_-_6328278 | 0.18 |
ENSDART00000130760
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr19_-_3115104 | 0.18 |
ENSDART00000093146
|
bop1
|
block of proliferation 1 |
| chr25_+_34410718 | 0.17 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr22_-_22277386 | 0.17 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr10_-_180681 | 0.16 |
|
|
|
| chr2_+_21698145 | 0.16 |
ENSDART00000145670
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr18_+_7584201 | 0.16 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr5_+_19703933 | 0.16 |
|
|
|
| chr20_-_25546095 | 0.15 |
ENSDART00000145034
|
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr5_+_30386272 | 0.15 |
ENSDART00000005893
|
ube2g1a
|
ubiquitin-conjugating enzyme E2G 1a (UBC7 homolog, yeast) |
| chr1_-_50438531 | 0.14 |
|
|
|
| chr3_-_26060748 | 0.14 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr12_+_27001284 | 0.14 |
|
|
|
| chr16_-_43329789 | 0.14 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr7_+_22710714 | 0.13 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr8_-_9531866 | 0.13 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr8_-_28359072 | 0.12 |
ENSDART00000062693
|
ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr11_+_30005670 | 0.12 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr12_+_26785950 | 0.12 |
ENSDART00000087329
|
znf438
|
zinc finger protein 438 |
| chr18_-_20469253 | 0.12 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr16_-_43329622 | 0.11 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr3_+_12438915 | 0.11 |
ENSDART00000159252
|
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr17_-_16414577 | 0.11 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr10_+_26910265 | 0.11 |
ENSDART00000166836
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr10_-_2761480 | 0.11 |
ENSDART00000131749
|
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr8_-_34509255 | 0.11 |
ENSDART00000112854
|
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr10_+_26910335 | 0.10 |
ENSDART00000166836
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr10_+_3052919 | 0.10 |
ENSDART00000028870
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr9_+_29832294 | 0.10 |
ENSDART00000023210
|
trim13
|
tripartite motif containing 13 |
| chr10_-_27023435 | 0.10 |
ENSDART00000050585
|
znhit2
|
zinc finger, HIT-type containing 2 |
| chr6_+_29396163 | 0.10 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr13_-_2211438 | 0.10 |
ENSDART00000137124
|
arfgef3
|
ARFGEF family member 3 |
| chr13_-_17938316 | 0.10 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr6_-_25065773 | 0.09 |
ENSDART00000165170
|
znf326
|
zinc finger protein 326 |
| chr10_+_28273581 | 0.08 |
ENSDART00000131003
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr1_+_31609119 | 0.08 |
ENSDART00000135078
|
pudp
|
pseudouridine 5'-phosphatase |
| chr3_-_24977167 | 0.08 |
ENSDART00000055432
|
rbx1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr23_-_16755682 | 0.08 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr18_+_23211146 | 0.07 |
|
|
|
| chr15_+_5913761 | 0.06 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr10_+_16111920 | 0.06 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr15_+_8791540 | 0.06 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr11_+_14046768 | 0.05 |
ENSDART00000059752
|
med16
|
mediator complex subunit 16 |
| chr7_-_67741663 | 0.05 |
|
|
|
| chr6_+_9558204 | 0.04 |
ENSDART00000108524
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr21_-_3619164 | 0.04 |
ENSDART00000133980
|
scamp1
|
secretory carrier membrane protein 1 |
| chr15_-_46484516 | 0.03 |
ENSDART00000154577
|
zgc:162872
|
zgc:162872 |
| chr19_-_26109474 | 0.03 |
ENSDART00000136187
|
bloc1s4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr10_+_42158472 | 0.03 |
ENSDART00000160687
|
setd1ba
|
SET domain containing 1B, a |
| chr10_+_3053045 | 0.03 |
ENSDART00000028870
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| KN149790v1_-_94139 | 0.02 |
ENSDART00000168319
|
zgc:171422
|
zgc:171422 |
| chr21_-_26973913 | 0.01 |
ENSDART00000147090
|
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.8 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.9 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |