Project
DANIO-CODE
Navigation
Downloads
Results for hoxd11a
Z-value: 0.63
Transcription factors associated with hoxd11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd11a
|
ENSDARG00000059267 | homeobox D11a |
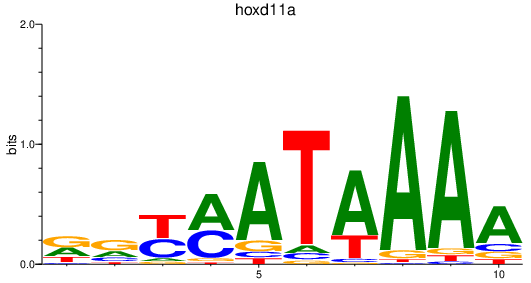
Activity profile of hoxd11a motif
Sorted Z-values of hoxd11a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd11a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_34150130 | 2.75 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr11_-_25019899 | 1.92 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr21_+_20360180 | 1.79 |
ENSDART00000003299
|
ENSDARG00000025174
|
ENSDARG00000025174 |
| chr15_+_29092022 | 1.46 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr5_-_23211957 | 1.43 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr19_-_868378 | 1.35 |
|
|
|
| chr8_-_2557556 | 1.23 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr3_+_44928323 | 1.13 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr6_+_28218420 | 1.12 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr22_-_22139268 | 1.10 |
ENSDART00000101659
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr17_+_14957568 | 1.05 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr8_-_2557506 | 1.04 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr20_-_34126039 | 1.04 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr3_+_44928592 | 0.99 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr2_+_23383492 | 0.98 |
ENSDART00000170669
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr6_+_36862078 | 0.96 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr15_+_29091983 | 0.96 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr5_-_31257158 | 0.94 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr5_-_35997345 | 0.94 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr9_+_22166405 | 0.93 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr23_+_22858063 | 0.92 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr15_-_19192862 | 0.90 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr9_+_33343936 | 0.89 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr20_-_40357302 | 0.88 |
|
|
|
| chr22_+_1837102 | 0.86 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr23_+_28563324 | 0.82 |
ENSDART00000006657
|
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr19_-_868837 | 0.82 |
|
|
|
| chr25_+_18855868 | 0.81 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr9_-_8916860 | 0.81 |
ENSDART00000138527
|
rab20
|
RAB20, member RAS oncogene family |
| chr5_-_29818204 | 0.81 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr9_+_33344121 | 0.81 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr9_+_22166482 | 0.78 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr12_+_33257120 | 0.78 |
|
|
|
| chr2_-_32755182 | 0.77 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr13_-_26668719 | 0.76 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr3_-_26052785 | 0.76 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr25_-_34468120 | 0.76 |
ENSDART00000175141
|
cdk10
|
cyclin-dependent kinase 10 |
| chr13_-_15662476 | 0.75 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr8_+_41003546 | 0.74 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr2_-_37116119 | 0.73 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr8_+_13327234 | 0.72 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr3_-_29779725 | 0.71 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr15_+_44160948 | 0.71 |
ENSDART00000110060
|
zgc:165514
|
zgc:165514 |
| chr13_-_15662599 | 0.69 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr13_-_37340209 | 0.67 |
|
|
|
| chr8_-_20882626 | 0.67 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr11_+_24076334 | 0.65 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr12_-_33257026 | 0.65 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr25_+_20596490 | 0.65 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr17_-_9794176 | 0.65 |
|
|
|
| chr22_-_22139099 | 0.64 |
|
|
|
| chr3_-_29779598 | 0.63 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr5_-_53907908 | 0.63 |
ENSDART00000158069
|
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr24_+_21575641 | 0.63 |
ENSDART00000137952
|
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr18_+_15695287 | 0.62 |
|
|
|
| chr10_-_25448712 | 0.62 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr10_-_43874597 | 0.61 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr11_-_25019298 | 0.61 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr23_+_22857958 | 0.61 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr17_+_17745119 | 0.60 |
ENSDART00000138189
ENSDART00000105013 |
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr3_-_30985152 | 0.60 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr22_+_3989571 | 0.60 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr8_-_38284959 | 0.59 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr23_+_43916520 | 0.59 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr19_+_9536231 | 0.57 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr24_+_21575038 | 0.57 |
ENSDART00000048273
|
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr8_-_38284748 | 0.57 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr5_-_29818325 | 0.56 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr6_-_42390995 | 0.56 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr6_+_36862148 | 0.56 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr2_-_44924505 | 0.56 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr2_+_35750334 | 0.53 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr3_-_26052601 | 0.53 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr8_-_39850462 | 0.53 |
ENSDART00000131372
|
mlec
|
malectin |
| chr23_-_43916621 | 0.53 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr21_-_38979792 | 0.53 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr20_+_23707789 | 0.52 |
|
|
|
| chr10_-_42931440 | 0.52 |
ENSDART00000123608
|
zgc:100918
|
zgc:100918 |
| chr5_-_35997304 | 0.51 |
ENSDART00000031270
|
rhogc
|
ras homolog gene family, member Gc |
| chr18_+_45866556 | 0.51 |
ENSDART00000024615
|
rnpepl1
|
arginyl aminopeptidase like 1 |
| chr20_+_25687135 | 0.51 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr16_-_28723759 | 0.50 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr2_+_23383257 | 0.50 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr14_-_30642610 | 0.49 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr3_+_39437497 | 0.49 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr8_-_18869085 | 0.48 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr3_+_52290252 | 0.48 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr6_+_49552893 | 0.47 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr15_+_22510807 | 0.47 |
ENSDART00000040542
|
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr1_-_8565788 | 0.46 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr5_-_31257031 | 0.46 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr10_-_43874646 | 0.46 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr3_+_39437613 | 0.46 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr9_+_19679106 | 0.46 |
ENSDART00000147148
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr23_-_39253291 | 0.45 |
|
|
|
| chr2_+_23383380 | 0.45 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr3_+_52290504 | 0.45 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr19_+_7630777 | 0.45 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr5_-_4483970 | 0.44 |
|
|
|
| chr12_-_33256934 | 0.44 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr15_+_44069851 | 0.44 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr20_-_36510718 | 0.43 |
|
|
|
| chr2_+_34675356 | 0.43 |
|
|
|
| chr10_-_11011134 | 0.42 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr19_-_869177 | 0.42 |
|
|
|
| chr24_-_32110851 | 0.41 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr20_+_52019524 | 0.41 |
ENSDART00000175674
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr16_-_21128542 | 0.41 |
ENSDART00000154833
|
CR392024.1
|
ENSDARG00000096565 |
| chr8_-_38284904 | 0.41 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr8_+_10824790 | 0.40 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr25_+_16259634 | 0.39 |
ENSDART00000136454
|
tead1a
|
TEA domain family member 1a |
| chr3_+_54507075 | 0.38 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr25_+_20596613 | 0.38 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr10_+_5234038 | 0.38 |
ENSDART00000063120
|
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr17_+_31205274 | 0.37 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr2_-_32754956 | 0.37 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr7_+_19300487 | 0.36 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr17_-_29295450 | 0.36 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr23_+_34120843 | 0.35 |
ENSDART00000143339
|
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr6_-_19794771 | 0.35 |
ENSDART00000151444
|
setd5
|
SET domain containing 5 |
| chr3_-_29779552 | 0.35 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr8_-_22492972 | 0.34 |
|
|
|
| chr4_-_16846903 | 0.34 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr23_+_43916552 | 0.34 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr3_+_48811662 | 0.33 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr11_+_24076371 | 0.32 |
ENSDART00000166045
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr20_-_2696401 | 0.32 |
ENSDART00000104606
|
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr14_-_30642549 | 0.32 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr17_+_8642428 | 0.31 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr14_+_38509671 | 0.30 |
|
|
|
| chr9_+_38778234 | 0.30 |
ENSDART00000131784
|
snx4
|
sorting nexin 4 |
| chr14_+_38510523 | 0.30 |
|
|
|
| chr10_+_45184723 | 0.29 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr19_+_42758938 | 0.28 |
ENSDART00000077059
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr8_+_26391153 | 0.26 |
ENSDART00000053447
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr3_-_30984999 | 0.26 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr7_+_65072867 | 0.26 |
ENSDART00000162693
|
bola3
|
bolA family member 3 |
| chr2_+_16928379 | 0.25 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr1_+_15928362 | 0.25 |
ENSDART00000149026
ENSDART00000159785 ENSDART00000164899 |
pcm1
|
pericentriolar material 1 |
| chr25_+_18855775 | 0.25 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr24_-_32110808 | 0.24 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr10_-_8238422 | 0.24 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr8_+_23710343 | 0.24 |
ENSDART00000166451
|
fance
|
Fanconi anemia, complementation group E |
| chr18_+_5260404 | 0.23 |
ENSDART00000150992
|
wdr59
|
WD repeat domain 59 |
| chr14_+_38510427 | 0.22 |
|
|
|
| chr21_+_3017623 | 0.22 |
|
|
|
| chr17_+_8642686 | 0.22 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr23_+_34120812 | 0.22 |
ENSDART00000143933
|
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr7_+_13386796 | 0.22 |
ENSDART00000157870
|
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr25_-_20160885 | 0.21 |
ENSDART00000160700
|
dnm1l
|
dynamin 1-like |
| chr8_+_41003629 | 0.20 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr19_+_7630609 | 0.19 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr6_+_36900159 | 0.19 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr16_-_28723722 | 0.19 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr5_+_44205128 | 0.18 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr17_+_23534824 | 0.18 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr3_-_40790889 | 0.18 |
|
|
|
| chr20_-_51454257 | 0.17 |
ENSDART00000023064
|
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr17_+_25269767 | 0.17 |
|
|
|
| chr25_-_7635613 | 0.16 |
ENSDART00000021577
|
phf21ab
|
PHD finger protein 21Ab |
| chr24_-_32603370 | 0.16 |
|
|
|
| chr20_+_51385348 | 0.15 |
|
|
|
| chr16_+_21128596 | 0.15 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr24_-_38758093 | 0.15 |
|
|
|
| chr8_-_38284846 | 0.15 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr8_-_38284660 | 0.15 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr5_+_50266345 | 0.14 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr16_-_6304889 | 0.14 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr24_+_21575343 | 0.12 |
ENSDART00000137952
|
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr2_+_50071638 | 0.12 |
ENSDART00000056248
|
wdr48b
|
WD repeat domain 48b |
| KN149711v1_-_7827 | 0.11 |
ENSDART00000171410
|
ENSDARG00000104905
|
ENSDARG00000104905 |
| chr24_-_36349914 | 0.10 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr3_-_40791035 | 0.10 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr9_+_3310926 | 0.09 |
ENSDART00000044128
|
hat1
|
histone acetyltransferase 1 |
| chr19_+_7630730 | 0.08 |
ENSDART00000115058
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr23_-_14647710 | 0.08 |
ENSDART00000090930
|
nkain4
|
sodium/potassium transporting ATPase interacting 4 |
| chr1_+_40758474 | 0.08 |
ENSDART00000139521
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr20_-_51454057 | 0.08 |
ENSDART00000023064
|
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr12_-_13611954 | 0.08 |
ENSDART00000124638
ENSDART00000124364 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr5_-_28167409 | 0.07 |
ENSDART00000158299
|
traf2a
|
Tnf receptor-associated factor 2a |
| chr9_+_13258490 | 0.07 |
ENSDART00000141705
|
carf
|
calcium responsive transcription factor |
| chr13_-_36738055 | 0.07 |
ENSDART00000177331
|
pygl
|
phosphorylase, glycogen, liver |
| chr24_-_11767965 | 0.07 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr25_-_6262259 | 0.06 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr6_-_40700033 | 0.06 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr21_-_3619164 | 0.06 |
ENSDART00000133980
|
scamp1
|
secretory carrier membrane protein 1 |
| chr24_+_11768279 | 0.06 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr5_+_66693263 | 0.05 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr6_-_6925121 | 0.05 |
ENSDART00000081760
|
ihhb
|
Indian hedgehog homolog b |
| chr18_+_38928197 | 0.04 |
|
|
|
| chr6_-_37767593 | 0.04 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr25_+_7359012 | 0.04 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr23_+_8862155 | 0.03 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr15_+_28249920 | 0.03 |
|
|
|
| chr10_+_15496976 | 0.03 |
ENSDART00000087680
|
erbb2ip
|
erbb2 interacting protein |
| chr23_-_36023579 | 0.03 |
|
|
|
| chr11_-_864648 | 0.03 |
ENSDART00000162152
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr3_+_24004141 | 0.02 |
ENSDART00000155216
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr19_-_33013748 | 0.02 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr7_-_18256590 | 0.02 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.5 | 1.4 | GO:0070841 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.3 | 1.7 | GO:0051754 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 0.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.3 | 0.9 | GO:0001659 | medium-chain fatty acid transport(GO:0001579) temperature homeostasis(GO:0001659) |
| 0.2 | 0.9 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.6 | GO:0035552 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 | 1.0 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 1.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.5 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.6 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.5 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.7 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 4.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.7 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.0 | 0.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0042113 | B cell activation(GO:0042113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 2.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 0.6 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 0.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 2.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.6 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.2 | 1.1 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 2.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 2.1 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |