Project
DANIO-CODE
Navigation
Downloads
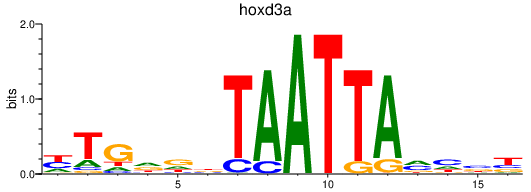
Results for hoxd3a
Z-value: 0.52
Transcription factors associated with hoxd3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd3a
|
ENSDARG00000059280 | homeobox D3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd3a | dr10_dc_chr9_-_1958795_1958837 | 0.88 | 8.0e-06 | Click! |
Activity profile of hoxd3a motif
Sorted Z-values of hoxd3a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd3a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_39141022 | 1.43 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr6_-_23193752 | 1.20 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr15_-_1858350 | 1.13 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr1_+_38834751 | 1.00 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr19_-_7069920 | 0.98 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr20_-_9107294 | 0.94 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr15_-_35394535 | 0.94 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr16_+_14817799 | 0.92 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr23_+_27749014 | 0.85 |
ENSDART00000128833
|
rps26
|
ribosomal protein S26 |
| chr12_-_27032151 | 0.78 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr23_-_7740845 | 0.77 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr10_+_39148251 | 0.71 |
ENSDART00000125986
|
ENSDARG00000055339
|
ENSDARG00000055339 |
| chr6_-_8075384 | 0.64 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr14_-_2682064 | 0.59 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr22_+_16282153 | 0.57 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr8_-_19166630 | 0.56 |
|
|
|
| chr10_-_26782374 | 0.54 |
ENSDART00000162710
|
fgf13b
|
fibroblast growth factor 13b |
| chr19_-_7069850 | 0.51 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr19_+_21783111 | 0.50 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr5_+_62987426 | 0.50 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr13_-_49695322 | 0.49 |
|
|
|
| chr1_-_50215233 | 0.49 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr21_+_8334453 | 0.48 |
ENSDART00000055336
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr23_+_36016450 | 0.46 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr17_-_37107310 | 0.46 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr7_+_9726412 | 0.46 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr7_+_25052687 | 0.45 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr23_+_17220363 | 0.45 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr3_+_27667194 | 0.43 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr7_-_67906887 | 0.39 |
ENSDART00000171943
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr6_-_57542101 | 0.39 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr8_+_17042976 | 0.38 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr20_+_29307142 | 0.35 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_-_44492897 | 0.34 |
|
|
|
| chr5_-_37494209 | 0.33 |
ENSDART00000084890
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr19_+_21783429 | 0.33 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr10_+_39140943 | 0.32 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr5_+_65754237 | 0.31 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr6_-_23193873 | 0.28 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr8_+_25015374 | 0.26 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr20_-_15029379 | 0.26 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr20_+_29307039 | 0.23 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_+_25015325 | 0.23 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr2_+_50873893 | 0.22 |
|
|
|
| chr11_+_13000957 | 0.22 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr20_-_46458839 | 0.20 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr5_+_27560610 | 0.18 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr20_+_26012369 | 0.17 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr11_-_40464213 | 0.16 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr2_-_28687065 | 0.16 |
ENSDART00000164657
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr15_-_5911817 | 0.15 |
|
|
|
| chr18_-_510172 | 0.13 |
ENSDART00000164374
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr6_+_3120600 | 0.13 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr7_-_64172974 | 0.09 |
ENSDART00000172619
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr9_+_41910191 | 0.08 |
|
|
|
| chr2_+_50873843 | 0.07 |
|
|
|
| chr8_-_22252973 | 0.07 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr11_-_40464253 | 0.06 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr10_-_41478085 | 0.06 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr15_-_5911548 | 0.05 |
|
|
|
| chr20_+_29307283 | 0.05 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr10_-_20545884 | 0.05 |
|
|
|
| chr21_-_32449627 | 0.03 |
|
|
|
| chr7_-_32624437 | 0.02 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr14_+_22294338 | 0.02 |
ENSDART00000167829
|
graf4b
|
GDNF family receptor alpha 4b |
| chr11_+_35790782 | 0.01 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.5 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 0.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.1 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.6 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.6 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.4 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.5 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |